Spinal muscular atrophy type I associated with a novel SMN1 splicing variant that disrupts the expression of the functional transcript
- PMID: 37799281
- PMCID: PMC10548546
- DOI: 10.3389/fneur.2023.1241195
Spinal muscular atrophy type I associated with a novel SMN1 splicing variant that disrupts the expression of the functional transcript
Abstract
Introduction: Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder caused by pathogenic variants in the SMN1 gene. The majority of SMA patients harbor a homozygous deletion of SMN1 exon 7 (95%). Heterozygosity for a conventional variant and a deletion is rare (5%) and not easily detected, due to the highly homologous SMN2 gene interference. SMN2 mainly produces a truncated non-functional protein (SMN-d7) instead of the full-length functional (SMN-FL). We hereby report a novel SMN1 splicing variant in an infant with severe SMA.
Methods: MLPA was used for SMN1/2 exon dosage determination. Sanger sequencing approaches and long-range PCR were employed to search for an SMN1 variant. Conventional and improved Real-time PCR assays were developed for the qualitative and quantitative SMN1/2 RNA analysis.
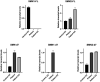
Results: The novel SMN1 splice-site variant c.835-8_835-5delinsG, was identified in compound heterozygosity with SMN1 exons 7/8 deletion. RNA studies revealed complete absence of SMN1 exon 7, thus confirming a disruptive effect of the variant on SMN1 splicing. No expression of the functional SMN1-FL transcript, remarkable expression of the SMN1-d7 and increased levels of the SMN2-FL/SMN2-d7 transcripts were observed.
Discussion: We verified the occurrence of a non-deletion SMN1 variant and supported its pathogenicity, thus expanding the SMN1 variants spectrum. We discuss the updated SMA genetic findings in the Cypriot population, highlighting an increased percentage of intragenic variants compared to other populations.
Keywords: Cypriot population; novel variant; spinal muscular atrophy; splicing dysregulation; transcript analysis.
Copyright © 2023 Votsi, Koutsou, Ververis, Georghiou, Nicolaou, Tanteles and Christodoulou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Revealing diverse alternative splicing variants of the highly homologous SMN1 and SMN2 genes by targeted long-read sequencing.Mol Genet Genomics. 2022 Jul;297(4):1039-1048. doi: 10.1007/s00438-022-01874-6. Epub 2022 May 25. Mol Genet Genomics. 2022. PMID: 35612622
-
The SMN1 common variant c.22 dupA in Chinese patients causes spinal muscular atrophy by nonsense-mediated mRNA decay in humans.Gene. 2018 Feb 20;644:49-55. doi: 10.1016/j.gene.2017.10.048. Epub 2017 Nov 20. Gene. 2018. PMID: 29080838
-
Dual Mechanism of a New SMN1 Variant (c.835G>C, p.Gly279Arg) by Interrupting Exon 7 Skipping and YG Oligomerization in Causation of Spinal Muscular Atrophy.J Mol Neurosci. 2021 Jan;71(1):112-121. doi: 10.1007/s12031-020-01631-7. Epub 2020 Aug 18. J Mol Neurosci. 2021. PMID: 32812185
-
Genetic testing and risk assessment for spinal muscular atrophy (SMA).Hum Genet. 2002 Dec;111(6):477-500. doi: 10.1007/s00439-002-0828-x. Epub 2002 Oct 3. Hum Genet. 2002. PMID: 12436240 Review.
-
An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA).Hum Mutat. 2000;15(3):228-37. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. Hum Mutat. 2000. PMID: 10679938 Review.
Cited by
-
In utero therapy for spinal muscular atrophy: closer to clinical translation.Brain. 2025 Sep 3;148(9):3043-3056. doi: 10.1093/brain/awaf123. Brain. 2025. PMID: 40193572 Free PMC article. Review.
-
In Search of Spinal Muscular Atrophy Disease Modifiers.Int J Mol Sci. 2024 Oct 18;25(20):11210. doi: 10.3390/ijms252011210. Int J Mol Sci. 2024. PMID: 39456991 Free PMC article. Review.
-
An Insightful Observation Leading to a Late Diagnosis of Spinal Muscular Atrophy: A Case Report.Cureus. 2024 May 7;16(5):e59786. doi: 10.7759/cureus.59786. eCollection 2024 May. Cureus. 2024. PMID: 38846202 Free PMC article.
References
LinkOut - more resources
Full Text Sources

