Small molecules targeting protein-protein interactions for cancer therapy
- PMID: 37799384
- PMCID: PMC10547922
- DOI: 10.1016/j.apsb.2023.05.035
Small molecules targeting protein-protein interactions for cancer therapy
Abstract
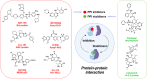
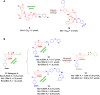
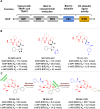
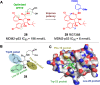
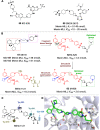
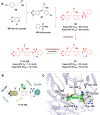
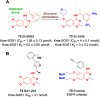
Protein-protein interactions (PPIs) are fundamental to many biological processes that play an important role in the occurrence and development of a variety of diseases. Targeting the interaction between tumour-related proteins with emerging small molecule drugs has become an attractive approach for treatment of human diseases, especially tumours. Encouragingly, selective PPI-based therapeutic agents have been rapidly advancing over the past decade, providing promising perspectives for novel therapies for patients with cancer. In this review we comprehensively clarify the discovery and development of small molecule modulators of PPIs from multiple aspects, focusing on PPIs in disease, drug design and discovery strategies, structure-activity relationships, inherent dilemmas, and future directions.
Keywords: Cancer; Protein‒protein interactions; Small molecules; Structure-activity relationships.
© 2023 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures













Similar articles
-
Emerging roles of allosteric modulators in the regulation of protein-protein interactions (PPIs): A new paradigm for PPI drug discovery.Med Res Rev. 2019 Nov;39(6):2314-2342. doi: 10.1002/med.21585. Epub 2019 Apr 7. Med Res Rev. 2019. PMID: 30957264 Review.
-
Methods for the Discovery and Identification of Small Molecules Targeting Oxidative Stress-Related Protein-Protein Interactions: An Update.Antioxidants (Basel). 2022 Mar 23;11(4):619. doi: 10.3390/antiox11040619. Antioxidants (Basel). 2022. PMID: 35453304 Free PMC article. Review.
-
Targeting protein-protein interactions in hematologic malignancies: still a challenge or a great opportunity for future therapies?Immunol Rev. 2015 Jan;263(1):279-301. doi: 10.1111/imr.12244. Immunol Rev. 2015. PMID: 25510283 Free PMC article. Review.
-
State-of-the-art strategies for targeting protein-protein interactions by small-molecule inhibitors.Chem Soc Rev. 2015 Nov 21;44(22):8238-59. doi: 10.1039/c5cs00252d. Epub 2015 Aug 6. Chem Soc Rev. 2015. PMID: 26248294 Review.
-
Development of Allosteric Modulators of Voltage-Gated Na+ Channels: A Novel Approach for an Old Target.Curr Top Med Chem. 2021;21(10):841-848. doi: 10.2174/1568026621666210525105359. Curr Top Med Chem. 2021. PMID: 34036922 Free PMC article. Review.
Cited by
-
Targeting autophagy and beyond: Deconvoluting the complexity of Beclin-1 from biological function to cancer therapy.Acta Pharm Sin B. 2023 Dec;13(12):4688-4714. doi: 10.1016/j.apsb.2023.08.008. Epub 2023 Aug 12. Acta Pharm Sin B. 2023. PMID: 38045051 Free PMC article. Review.
-
Beyond ribosomal function: RPS6 deficiency suppresses cholangiocarcinoma cell growth by disrupting alternative splicing.Acta Pharm Sin B. 2024 Sep;14(9):3931-3948. doi: 10.1016/j.apsb.2024.06.028. Epub 2024 Jun 29. Acta Pharm Sin B. 2024. PMID: 39309509 Free PMC article.
-
DeepTGIN: a novel hybrid multimodal approach using transformers and graph isomorphism networks for protein-ligand binding affinity prediction.J Cheminform. 2024 Dec 29;16(1):147. doi: 10.1186/s13321-024-00938-6. J Cheminform. 2024. PMID: 39734235 Free PMC article.
-
Recent advances in Alzheimer's disease: Mechanisms, clinical trials and new drug development strategies.Signal Transduct Target Ther. 2024 Aug 23;9(1):211. doi: 10.1038/s41392-024-01911-3. Signal Transduct Target Ther. 2024. PMID: 39174535 Free PMC article. Review.
-
The permissive binding theory of cancer.Front Oncol. 2023 Nov 9;13:1272981. doi: 10.3389/fonc.2023.1272981. eCollection 2023. Front Oncol. 2023. PMID: 38023252 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

