Antibody-drug conjugates: Recent advances in payloads
- PMID: 37799390
- PMCID: PMC10547921
- DOI: 10.1016/j.apsb.2023.06.015
Antibody-drug conjugates: Recent advances in payloads
Abstract
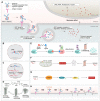
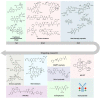
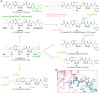
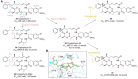
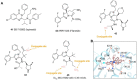
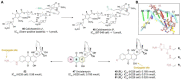
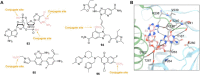
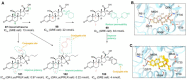
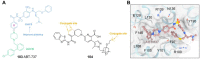
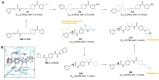
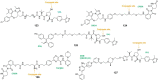
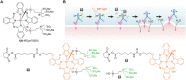
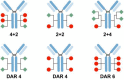
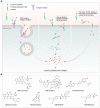
Antibody‒drug conjugates (ADCs), which combine the advantages of monoclonal antibodies with precise targeting and payloads with efficient killing, show great clinical therapeutic value. The ADCs' payloads play a key role in determining the efficacy of ADC drugs and thus have attracted great attention in the field. An ideal ADC payload should possess sufficient toxicity, low immunogenicity, high stability, and modifiable functional groups. Common ADC payloads include tubulin inhibitors and DNA damaging agents, with tubulin inhibitors accounting for more than half of the ADC drugs in clinical development. However, due to clinical limitations of traditional ADC payloads, such as inadequate efficacy and the development of acquired drug resistance, novel highly efficient payloads with diverse targets and reduced side effects are being developed. This perspective summarizes the recent research advances of traditional and novel ADC payloads with main focuses on the structure-activity relationship studies, co-crystal structures, and designing strategies, and further discusses the future research directions of ADC payloads. This review also aims to provide valuable references and future directions for the development of novel ADC payloads that will have high efficacy, low toxicity, adequate stability, and abilities to overcome drug resistance.
Keywords: Antibody‒drug conjugates; DNA damaging agents; Dual payloads; PROTACs; RNA targeting agents; Tubulin inhibitors.
© 2023 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures










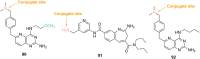

References
-
- Weingart S.N., Zhang L., Sweeney M., Hassett M. Chemotherapy medication errors. Lancet Oncol. 2018;19:e191–e199. - PubMed
-
- Zraik I.M., Hess-Busch Y. Management of chemotherapy side effects and their long-term sequelae. Urologe. 2021;60:862–871. - PubMed
-
- Chau C.H., Steeg P.S., Figg W.D. Antibody‒drug conjugates for cancer. Lancet. 2019;394:793–804. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

