Full-length transcriptome analysis revealed that 2,4-dichlorophenoxyacetic acid promoted in vitro bulblet initiation in lily by affecting carbohydrate metabolism and auxin signaling
- PMID: 37799550
- PMCID: PMC10548195
- DOI: 10.3389/fpls.2023.1236315
Full-length transcriptome analysis revealed that 2,4-dichlorophenoxyacetic acid promoted in vitro bulblet initiation in lily by affecting carbohydrate metabolism and auxin signaling
Abstract
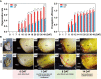
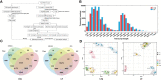
Bulblet initiation, including adventitious bud initiation and bulblet formation, is a crucial process for lily and other bulbous flowers that are commercially propagated by vegetative means. Here, by a hybrid strategy combining Pacific Biosciences (PacBio) full-length sequencing and Illumina RNA sequencing (RNA-seq), high-quality transcripts of L. brownii (Lb) and its variety, L. brownii var. giganteum (Lbg), during in vitro bulblet initiation were obtained. A total of 53,576 and 65,050 high-quality non-redundant full-length transcripts of Lbg and Lb were generated, respectively. Morphological observation showed that Lbg possessed a stronger capacity to generate bulblets in vitro than Lb, and 1 mg L-1 2,4-dichlorophenoxyacetic acid (2,4-D) significantly increased bulblet regeneration rate in two lilies. Screening of differentially expressed transcripts (DETs) between different stages and Mfuzz analysis showed 0 DAT to 1 DAT was the crucial stage with the most complex transcriptional change, with carbohydrate metabolism pathway was significantly enriched. In addition, 6,218 and 8,965 DETs were screened between the 2,4-D-treated group and the control group in Lbg and Lb, respectively. 2,4-D application had evident effects on the expression of genes involved in auxin signaling pathway, such as TIRs, ARFs, Aux/IAAs, GH3s and SAURs. Then, we compared the expression profiles of crucial genes of carbohydrate metabolism between different stages and different treatments. SUSs, SUTs, TPSs, AGPLs, GBSSs and SSs showed significant responses during bulblet initiation. The expression of CWINs, SUTs and SWEETs were significantly upregulated by 2,4-D in two lilies. In addition, 2,4-D increased the expression of starch degradation genes (AMYs and BAMs) and inhibited starch synthesis genes (AGPLs, GBSSs and SSs). SBEs were significantly upregulated in Lbg but not in Lb. Significant co-expression was showed between genes involved in carbohydrate metabolism and auxin signaling, together with transcription factors such as bHLHs, MYBs, ERFs and C3Hs. This study indicates the coordinate regulation of bulblet initiation by carbohydrate metabolism and auxin signaling, serving as a basis for further studies on the molecular mechanism of bulblet initiation in lily and other bulbous flowers.
Keywords: Lilium; auxin signaling; full-length sequencing; starch synthesis and degradation; sucrose unloading.
Copyright © 2023 Gao, Zhang, Xu, Liu, Xiao, Cui, Xia, Wu and Ren.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor.BMC Plant Biol. 2014 Dec 19;14:358. doi: 10.1186/s12870-014-0358-4. BMC Plant Biol. 2014. PMID: 25524032 Free PMC article.
-
Hydrogen-rich water promotes the formation of bulblets in Lilium davidii var. unicolor through regulating sucrose and starch metabolism.Planta. 2021 Oct 24;254(5):106. doi: 10.1007/s00425-021-03762-6. Planta. 2021. PMID: 34689230 Free PMC article.
-
Low humic acids promote in vitro lily bulblet enlargement by enhancing roots growth and carbohydrate metabolism.J Zhejiang Univ Sci B. 2016 Nov.;17(11):892-904. doi: 10.1631/jzus.B1600231. J Zhejiang Univ Sci B. 2016. PMID: 27819136 Free PMC article.
-
Transcriptome analysis of starch and sucrose metabolism across bulb development in Sagittaria sagittifolia.Gene. 2018 Apr 5;649:99-112. doi: 10.1016/j.gene.2018.01.075. Epub 2018 Jan 31. Gene. 2018. PMID: 29374598 Review.
-
Auxin response factors are keys to the many auxin doors.New Phytol. 2022 Jul;235(2):402-419. doi: 10.1111/nph.18159. Epub 2022 May 10. New Phytol. 2022. PMID: 35434800 Review.
Cited by
-
New links between auxin and starch.Nat Commun. 2025 Jan 19;16(1):491. doi: 10.1038/s41467-024-55756-z. Nat Commun. 2025. PMID: 39828730 Free PMC article.
-
Identification and Expression Analysis of Putative Sugar Transporter Gene Family during Bulb Formation in Lilies.Int J Mol Sci. 2024 Mar 20;25(6):3483. doi: 10.3390/ijms25063483. Int J Mol Sci. 2024. PMID: 38542476 Free PMC article.
References
LinkOut - more resources
Full Text Sources

