Unveiling the regulatory mechanisms of salicylate degradation gene cluster cehGHIR4 in Rhizobium sp. strain X9
- PMID: 37800922
- PMCID: PMC10617420
- DOI: 10.1128/aem.00802-23
Unveiling the regulatory mechanisms of salicylate degradation gene cluster cehGHIR4 in Rhizobium sp. strain X9
Abstract
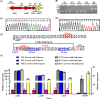
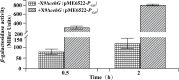
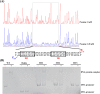
In a previous study, the novel gene cluster cehGHI was found to be involved in salicylate degradation through the CoA-mediated pathway in Rhizobium sp. strain X9 (Mol Microbiol 116:783-793, 2021). In this study, an IclR family transcriptional regulator CehR4 was identified. In contrast to other regulators involved in salicylate degradation, cehR4 forms one operon with the gentisyl-CoA thioesterase gene cehI, while cehG and cehH (encoding salicylyl-CoA ligase and salicylyl-CoA hydroxylase, respectively) form another operon. cehGH and cehIR4 are divergently transcribed, and their promoters overlap. The results of the electrophoretic mobility shift assay and DNase I footprinting showed that CehR4 binds to the 42-bp motif between genes cehH and cehI, thus regulating transcription of cehGH and cehIR4. The repeat sequences IR1 (5'-TTTATATAAA-3') and IR2 (5'-AATATAGAAA-3') in the motif are key sites for CehR4 binding. The arrangement of cehGH and cehIR4 and the conserved binding motif of CehR4 were also found in other bacterial genera. The results disclose the regulatory mechanism of salicylate degradation through the CoA pathway and expand knowledge about the systems controlled by IclR family transcriptional regulators.IMPORTANCEThe long-term residue of aromatic compounds in the environment has brought great threat to the environment and human health. Microbial degradation plays an important role in the elimination of aromatic compounds in the environment. Salicylate is a common intermediate metabolite in the degradation of various aromatic compounds. Recently, Rhizobium sp. strain X9, capable of degrading the pesticide carbaryl, was isolated from carbaryl-contaminated soil. Salicylate is the intermediate metabolite that appeared during the degradation of carbaryl, and a novel salicylate degradation pathway and the involved gene cluster cehGHIR4 have been identified. This study identified and characterized the IclR transcription regulator CehR4 that represses transcription of cehGHIR4 gene cluster. Additionally, the genetic arrangements of cehGH and cehIR4 and the binding sites of CehR4 were also found in other bacterial genera. This study provides insights into the biodegradation of salicylate and provides an application in the bioremediation of aromatic compound-contaminated environments.
Keywords: IclR-type transcriptional regulators; Rhizobium sp. strain X9; cehR4; salicylate degradation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

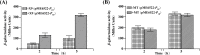

Similar articles
-
Unveiling the CoA mediated salicylate catabolic mechanism in Rhizobium sp. X9.Mol Microbiol. 2021 Sep;116(3):783-793. doi: 10.1111/mmi.14771. Epub 2021 Jun 28. Mol Microbiol. 2021. PMID: 34121246
-
Two LysR Family Transcriptional Regulators, McbH and McbN, Activate the Operons Responsible for the Midstream and Downstream Pathways, Respectively, of Carbaryl Degradation in Pseudomonas sp. Strain XWY-1.Appl Environ Microbiol. 2022 Feb 22;88(4):e0206021. doi: 10.1128/AEM.02060-21. Epub 2021 Dec 22. Appl Environ Microbiol. 2022. PMID: 34936841 Free PMC article.
-
McbG, a LysR Family Transcriptional Regulator, Activates the mcbBCDEF Gene Cluster Involved in the Upstream Pathway of Carbaryl Degradation in Pseudomonas sp. Strain XWY-1.Appl Environ Microbiol. 2021 Apr 13;87(9):e02970-20. doi: 10.1128/AEM.02970-20. Print 2021 Apr 13. Appl Environ Microbiol. 2021. PMID: 33579686 Free PMC article.
-
Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors.FEMS Microbiol Rev. 2006 Mar;30(2):157-86. doi: 10.1111/j.1574-6976.2005.00008.x. FEMS Microbiol Rev. 2006. PMID: 16472303 Review.
-
Bacterial transcriptional regulators for degradation pathways of aromatic compounds.Microbiol Mol Biol Rev. 2004 Sep;68(3):474-500, table of contents. doi: 10.1128/MMBR.68.3.474-500.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353566 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

