Mechanical stress confers nuclear and functional changes in derived leukemia cells from persistent confined migration
- PMID: 37801090
- PMCID: PMC10558412
- DOI: 10.1007/s00018-023-04968-5
Mechanical stress confers nuclear and functional changes in derived leukemia cells from persistent confined migration
Abstract
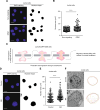
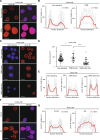
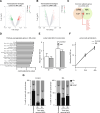
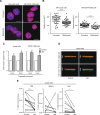
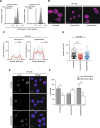
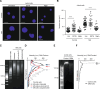
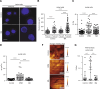
Nuclear deformability plays a critical role in cell migration. During this process, the remodeling of internal components of the nucleus has a direct impact on DNA damage and cell behavior; however, how persistent migration promotes nuclear changes leading to phenotypical and functional consequences remains poorly understood. Here, we described that the persistent migration through physical barriers was sufficient to promote permanent modifications in migratory-altered cells. We found that derived cells from confined migration showed changes in lamin B1 localization, cell morphology and transcription. Further analysis confirmed that migratory-altered cells showed functional differences in DNA repair, cell response to chemotherapy and cell migration in vivo homing experiments. Experimental modulation of actin polymerization affected the redistribution of lamin B1, and the basal levels of DNA damage in migratory-altered cells. Finally, since major nuclear changes were present in migratory-altered cells, we applied a multidisciplinary biochemical and biophysical approach to identify that confined conditions promoted a different biomechanical response of the nucleus in migratory-altered cells. Our observations suggest that mechanical compression during persistent cell migration has a role in stable nuclear and genomic alterations that might handle the genetic instability and cellular heterogeneity in aging diseases and cancer.
Keywords: Cell migration; DNA damage; Lamin; Mechanobiology; Nuclear deformability.
© 2023. The Author(s).
Conflict of interest statement
Authors declare no competing interests.
Figures

References
MeSH terms
Grants and funding
- SAF2017-86327-R/Agencia Estatal de Investigación
- PDI2020-118525RB-I00/Agencia Estatal de Investigación
- RTI2018-097267-B-I00/Agencia Estatal de Investigación
- FPI Scholarship 2018/Agencia Estatal de Investigación
- PID2020-115444GB-I00/Agencia Estatal de Investigación
- PID2019-108391RB-100/Agencia Estatal de Investigación
- TED2021-132296B-C52/Agencia Estatal de Investigación
- Beca Leonardo 2020/Fundación BBVA
- PEJ-2020-AI/BMD-19152/Consejería de Educación, Juventud y Deporte, Comunidad de Madrid
- Y2018/BIO-5207/Consejería de Educación, Juventud y Deporte, Comunidad de Madrid
- Y2018/BIO-5207/Consejería de Educación, Juventud y Deporte, Comunidad de Madrid
- Y2018/BIO-5207/Consejería de Educación, Juventud y Deporte, Comunidad de Madrid
- BFERO2021.01/Fundación Fero
- LABAE211656TORR/Fundación Científica Asociación Española Contra el Cáncer
- S2018/NMT-4389/Comunidad de Madrid
- REACT-EU program PR38-21-28 ANTICIPA-CM/Comunidad de Madrid
- JAE Intro 2022/Fundación General CSIC
- JAE Intro 2022/Fundación General CSIC
LinkOut - more resources
Full Text Sources
Medical
Research Materials

