S-acylation of p62 promotes p62 droplet recruitment into autophagosomes in mammalian autophagy
- PMID: 37802024
- PMCID: PMC10552648
- DOI: 10.1016/j.molcel.2023.09.004
S-acylation of p62 promotes p62 droplet recruitment into autophagosomes in mammalian autophagy
Abstract
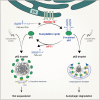
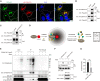
p62 is a well-characterized autophagy receptor that recognizes and sequesters specific cargoes into autophagosomes for degradation. p62 promotes the assembly and removal of ubiquitinated proteins by forming p62-liquid droplets. However, it remains unclear how autophagosomes efficiently sequester p62 droplets. Herein, we report that p62 undergoes reversible S-acylation in multiple human-, rat-, and mouse-derived cell lines, catalyzed by zinc-finger Asp-His-His-Cys S-acyltransferase 19 (ZDHHC19) and deacylated by acyl protein thioesterase 1 (APT1). S-acylation of p62 enhances the affinity of p62 for microtubule-associated protein 1 light chain 3 (LC3)-positive membranes and promotes autophagic membrane localization of p62 droplets, thereby leading to the production of small LC3-positive p62 droplets and efficient autophagic degradation of p62-cargo complexes. Specifically, increasing p62 acylation by upregulating ZDHHC19 or by genetic knockout of APT1 accelerates p62 degradation and p62-mediated autophagic clearance of ubiquitinated proteins. Thus, the protein S-acylation-deacylation cycle regulates p62 droplet recruitment to the autophagic membrane and selective autophagic flux, thereby contributing to the control of selective autophagic clearance of ubiquitinated proteins.
Keywords: APT1; S-acylation; ZDHHC19; autophagy; autophagy receptor; liquid-liquid phase separation; p62 droplet; p62 protein; protein posttranslational modification; selective autophagy.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.W.T. is a founder and shareholder in Myricx Pharma Ltd. and receives consultancy or research funding from Kura Oncology, Pfizer Ltd., Samsara Therapeutics, Myricx Pharma Ltd., MSD, Exscientia, and Daiichi Sankyo.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

