TIM-3 blockade in diffuse intrinsic pontine glioma models promotes tumor regression and antitumor immune memory
- PMID: 37802053
- PMCID: PMC10644900
- DOI: 10.1016/j.ccell.2023.09.001
TIM-3 blockade in diffuse intrinsic pontine glioma models promotes tumor regression and antitumor immune memory
Abstract
Diffuse intrinsic pontine glioma (DIPG) is an aggressive brain stem tumor and the leading cause of pediatric cancer-related death. To date, these tumors remain incurable, underscoring the need for efficacious therapies. In this study, we demonstrate that the immune checkpoint TIM-3 (HAVCR2) is highly expressed in both tumor cells and microenvironmental cells, mainly microglia and macrophages, in DIPG. We show that inhibition of TIM-3 in syngeneic models of DIPG prolongs survival and produces long-term survivors free of disease that harbor immune memory. This antitumor effect is driven by the direct effect of TIM-3 inhibition in tumor cells, the coordinated action of several immune cell populations, and the secretion of chemokines/cytokines that create a proinflammatory tumor microenvironment favoring a potent antitumor immune response. This work uncovers TIM-3 as a bona fide target in DIPG and supports its clinical translation.
Keywords: DIPG; DMGs; TIM-3; diffuse midline glioma; immune checkpoint; immunotherapy; macrophages; microglia; pediatric brain tumor; tumor microenvironment.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.C.A. is a member of the SAB for Tizona Therapeutics, Trishula Therapeutics, Compass Therapeutics, Zumutor Biologics, ImmuneOncia, and Excepgen, which have interests in cancer immunotherapy. A.C.A. is a paid consultant for iTeos Therapeutics and Larkspur Biosciences. A.C.A.’s interests were reviewed and managed by the Brigham and Women’s Hospital. The rest of authors do not have potential conflicts of interest to disclose.
Figures

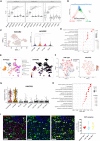
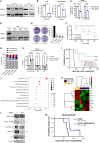


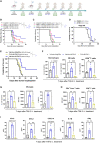

Comment in
-
The one-two punch: TIM-3 blockade targets immune and tumor cells to knock out pediatric brain tumors.Cancer Cell. 2023 Nov 13;41(11):1843-1845. doi: 10.1016/j.ccell.2023.09.016. Epub 2023 Oct 19. Cancer Cell. 2023. PMID: 37863067
References
-
- Hoffman L.M., Veldhuijzen van Zanten S.E.M., Colditz N., Baugh J., Chaney B., Hoffmann M., Lane A., Fuller C., Miles L., Hawkins C., et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of Diffuse Intrinsic Pontine Glioma (DIPG): A collaborative report from the International and European Society for Pediatric Oncology DIPG registries. J. Clin. Oncol. 2018;36:1963–1972. - PMC - PubMed
-
- El-Khouly F.E., Veldhuijzen van Zanten S.E.M., Santa-Maria Lopez V., Hendrikse N.H., Kaspers G.J.L., Loizos G., Sumerauer D., Nysom K., Pruunsild K., Pentikainen V., et al. Diagnostics and treatment of diffuse intrinsic pontine glioma: where do we stand? J. Neuro Oncol. 2019;145:177–184. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

