Human model of primary carnitine deficiency cardiomyopathy reveals ferroptosis as a novel mechanism
- PMID: 37802072
- PMCID: PMC10679537
- DOI: 10.1016/j.stemcr.2023.09.002
Human model of primary carnitine deficiency cardiomyopathy reveals ferroptosis as a novel mechanism
Abstract
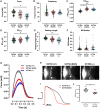
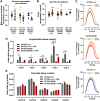
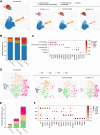
Primary carnitine deficiency (PCD) is an autosomal recessive monogenic disorder caused by mutations in SLC22A5. This gene encodes for OCTN2, which transports the essential metabolite carnitine into the cell. PCD patients suffer from muscular weakness and dilated cardiomyopathy. Two OCTN2-defective human induced pluripotent stem cell lines were generated, carrying a full OCTN2 knockout and a homozygous OCTN2 (N32S) loss-of-function mutation. OCTN2-defective genotypes showed lower force development and resting length in engineered heart tissue format compared with isogenic control. Force was sensitive to fatty acid-based media and associated with lipid accumulation, mitochondrial alteration, higher glucose uptake, and metabolic remodeling, replicating findings in animal models. The concordant results of OCTN2 (N32S) and -knockout emphasizes the relevance of OCTN2 for these findings. Importantly, genome-wide analysis and pharmacological inhibitor experiments identified ferroptosis, an iron- and lipid-dependent cell death pathway associated with fibroblast activation as a novel PCD cardiomyopathy disease mechanism.
Keywords: Cardiomyocytes; Dilated Cardiomyopathy; Metabolism; disease modeling; iPSC.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.E. is a member of the DiNAQOR Scientific Advisory Board and holds shares in DiNAQOR.
Figures





References
-
- Alesci S., Manoli I., Costello R., Coates P., Gold P.W., Chrousos G.P., Blackman M.R. Carnitine: Lessons from one hundred years of research. Ann. N. Y. Acad. Sci. 2004;1033
-
- Asai T., Okumura K., Takahashi R., Matsui H., Numaguchi Y., Murakami H., Murakami R., Murohara T. Combined therapy with PPARα agonist and l-carnitine rescues lipotoxic cardiomyopathy due to systemic carnitine deficiency. Cardiovasc. Res. 2006;70:566–577. - PubMed
-
- Bandet C.L., Hajduch E. CERT-Dependent Ceramide Transport, A Crucial Process in Cells. J Diabetes Clin Res. 2021;3:40–45.
-
- Breckwoldt K., Letuffe-Brenière D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S., et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. - PubMed
-
- Broderick T.L., Panagakis G., DiDomenico D., Gamble J., Lopaschuk G.D., Shug A.L., Paulson D.J. L-carnitine improvement of cardiac function is associated with a stimulation in glucose but not fatty acid metabolism in carnitine-deficient hearts. Cardiovasc. Res. 1995;30:815–820. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- NC/C013202/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- NC/T2T0119/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- PG/21/10545/BHF_/British Heart Foundation/United Kingdom
- SP/15/9/31605/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

