Advancing protein therapeutics through proximity-induced chemistry
- PMID: 37802076
- PMCID: PMC10960704
- DOI: 10.1016/j.chembiol.2023.09.004
Advancing protein therapeutics through proximity-induced chemistry
Abstract
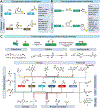
Recent years have seen a remarkable growth in the field of protein-based medical treatments. Nevertheless, concerns have arisen regarding the cytotoxicity limitations, low affinity, potential immunogenicity, low stability, and challenges to modify these proteins. To overcome these obstacles, proximity-induced chemistry has emerged as a next-generation strategy for advancing protein therapeutics. This method allows site-specific modification of proteins with therapeutic agents, improving their effectiveness without extensive engineering. In addition, this innovative approach enables spatial control of the reaction based on proximity, facilitating the formation of irreversible covalent bonds between therapeutic proteins and their targets. This capability becomes particularly valuable in addressing challenges such as the low affinity frequently encountered between therapeutic proteins and their targets, as well as the limited availability of small molecules for specific protein targets. As a result, proximity-induced chemistry is reshaping the field of protein drug preparation and propelling the revolution in novel protein therapeutics.
Keywords: covalent drug; cross-linking; protein drug; protein therapeutics; proximity-induced chemistry.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

