tRNA renovatio: Rebirth through fragmentation
- PMID: 37802077
- PMCID: PMC10841463
- DOI: 10.1016/j.molcel.2023.09.016
tRNA renovatio: Rebirth through fragmentation
Abstract
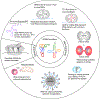
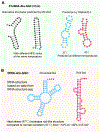
tRNA function is based on unique structures that enable mRNA decoding using anticodon trinucleotides. These structures interact with specific aminoacyl-tRNA synthetases and ribosomes using 3D shape and sequence signatures. Beyond translation, tRNAs serve as versatile signaling molecules interacting with other RNAs and proteins. Through evolutionary processes, tRNA fragmentation emerges as not merely random degradation but an act of recreation, generating specific shorter molecules called tRNA-derived small RNAs (tsRNAs). These tsRNAs exploit their linear sequences and newly arranged 3D structures for unexpected biological functions, epitomizing the tRNA "renovatio" (from Latin, meaning renewal, renovation, and rebirth). Emerging methods to uncover full tRNA/tsRNA sequences and modifications, combined with techniques to study RNA structures and to integrate AI-powered predictions, will enable comprehensive investigations of tRNA fragmentation products and new interaction potentials in relation to their biological functions. We anticipate that these directions will herald a new era for understanding biological complexity and advancing pharmaceutical engineering.
Keywords: AI-based RNA structure prediction; RNA structure; biological complexity; deep learning; evolution; non-canonical tRNA functions; tRNA; tRNA fragments; tRNA-derived small RNA.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Crick FH (1958). On protein synthesis. Symp Soc Exp Biol 12, 138–163. - PubMed
-
- Crick FH (1966). The genetic code--yesterday, today, and tomorrow. Cold Spring Harb Symp Quant Biol 31, 1–9. - PubMed
-
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, and Zamecnik PC (1958). A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231, 241–257. - PubMed
-
- Hoagland MB, Keller EB, and Zamecnik PC (1956). Enzymatic carboxyl activation of amino acids. J Biol Chem 218, 345–358. - PubMed
-
- Schimmel PR, and Soll D. (1979). Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem 48, 601–648. - PubMed

