Novel insights into the whole-blood DNA methylome of asthma in ethnically diverse children and youth
- PMID: 37802634
- PMCID: PMC10841414
- DOI: 10.1183/13993003.00714-2023
Novel insights into the whole-blood DNA methylome of asthma in ethnically diverse children and youth
Abstract
Background: The epigenetic mechanisms of asthma remain largely understudied in African Americans and Hispanics/Latinos, two populations disproportionately affected by asthma. We aimed to identify markers, regions and processes with differential patterns of DNA methylation (DNAm) in whole blood by asthma status in ethnically diverse children and youth, and to assess their functional consequences.
Methods: DNAm levels were profiled with the Infinium MethylationEPIC or HumanMethylation450 BeadChip arrays among 1226 African Americans or Hispanics/Latinos and assessed for differential methylation per asthma status at the CpG and region (differentially methylated region (DMR)) level. Novel associations were validated in blood and/or nasal epithelium from ethnically diverse children and youth. The functional and biological implications of the markers identified were investigated by combining epigenomics with transcriptomics from study participants.
Results: 128 CpGs and 196 DMRs were differentially methylated after multiple testing corrections, including 92.3% and 92.8% novel associations, respectively. 41 CpGs were replicated in other Hispanics/Latinos, prioritising cg17647904 (NCOR2) and cg16412914 (AXIN1) as asthma DNAm markers. Significant DNAm markers were enriched in previous associations for asthma, fractional exhaled nitric oxide, bacterial infections, immune regulation or eosinophilia. Functional annotation highlighted epigenetically regulated gene networks involved in corticosteroid response, host defence and immune regulation. Several implicated genes are targets for approved or experimental drugs, including TNNC1 and NDUFA12. Many differentially methylated loci previously associated with asthma were validated in our study.
Conclusions: We report novel whole-blood DNAm markers for asthma underlying key processes of the disease pathophysiology and confirm the transferability of previous asthma DNAm associations to ethnically diverse populations.
Copyright ©The authors 2023. For reproduction rights and permissions contact permissions@ersnet.org.
Conflict of interest statement
Conflict of interest: E. Herrera-Luis was supported by a fellowship awarded by MCIN/AEI/10.13039/501100011033 and by the European Social Fund (ESF) “Investing in your future” (PRE2018-083837). C. Rosa-Baez reports support for the present manuscript from the Ministry of Education and Vocational Training. M. Pino-Yanes was funded by the Ramón y Cajal Program (RYC-2015-17205) by MCIN/AEI/10.13039/501100011033 and by the ESF “Investing in your future”. She reports a grant from MCIN/AEI/10.13039/501100011033, and grant support from the EAACI Allergopharma Award 2021 and from GlaxoSmithKline, Spain, paid to Fundación Canaria Instituto de Investigación Sanitaria de Canarias for a project outside the submitted work. J. Villar and M. Pino-Yanes were also supported by CIBERES, Instituto de Salud Carlos III (ISCIII), Ministerio de Ciencia e Innovación and Unión Europea–European Regional Development Fund (CB06/06/1088). J. Villar also reports funding by ISCIII and the European Regional Development Fund “A way of making Europe”. E.G. Burchard reports grants from the National Institutes of Health, the Tobacco-Related Disease Research Program, the Sandler Family Foundation, the American Asthma Foundation, the Amos Medical Faculty Development Program from the Robert Wood Johnson Foundation, and from the Harry Wm. and Diana V. Hind Distinguished Professorship in Pharmaceutical Sciences II. C. Laprise is the director of the Centre Intersectoriel en Santé Durable, codirector of the Environment, Genetics and Cancer axis of the Respiratory Health Network, investigator of CHILD Study, member of the InFAC consortium, and chairholder of the Canada Research Chair in Asthma and Allergic Diseases Genomics. The remaining authors declare they have no competing interests or other interests that might be perceived to influence the interpretation of the article.
Figures



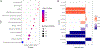
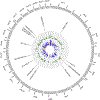

Comment in
-
Epigenome-wide association studies: the exposures of yesterday form the methylations of tomorrow.Eur Respir J. 2023 Dec 21;62(6):2301552. doi: 10.1183/13993003.01552-2023. Print 2023 Dec. Eur Respir J. 2023. PMID: 38128955 No abstract available.
References
-
- Centers for Disease Control and Prevention. National surveillance of asthma: United States, 2016–2018 [Internet]. Available at http://www.cdc.gov/asthma. Date last updated: September 22, 2022. Date last accessed: September 2019.
-
- Hernandez-Pacheco N, Flores C, Oh SS, et al. What Ancestry Can Tell Us About the Genetic Origins of Inter-Ethnic Differences in Asthma Expression. Curr Allergy Asthma Rep. 2016; 16: 53. - PubMed
-
- Benincasa G, DeMeo DL, Glass K, et al. Epigenetics and pulmonary diseases in the horizon of precision medicine: a review. Eur Respir J. 2021; 57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- UM1 HG008901/HG/NHGRI NIH HHS/United States
- R01 HL135156/HL/NHLBI NIH HHS/United States
- R01 HL128439/HL/NHLBI NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- R21 ES024844/ES/NIEHS NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- R56 MD013312/MD/NIMHD NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States
- HHSN268201600032C/ES/NIEHS NIH HHS/United States
- R01 HL155024/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
