Evaluation of a prototype machine learning tool to semi-automate data extraction for systematic literature reviews
- PMID: 37803451
- PMCID: PMC10557215
- DOI: 10.1186/s13643-023-02351-w
Evaluation of a prototype machine learning tool to semi-automate data extraction for systematic literature reviews
Abstract
Background: Evidence-based medicine requires synthesis of research through rigorous and time-intensive systematic literature reviews (SLRs), with significant resource expenditure for data extraction from scientific publications. Machine learning may enable the timely completion of SLRs and reduce errors by automating data identification and extraction.
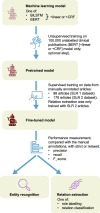
Methods: We evaluated the use of machine learning to extract data from publications related to SLRs in oncology (SLR 1) and Fabry disease (SLR 2). SLR 1 predominantly contained interventional studies and SLR 2 observational studies. Predefined key terms and data were manually annotated to train and test bidirectional encoder representations from transformers (BERT) and bidirectional long-short-term memory machine learning models. Using human annotation as a reference, we assessed the ability of the models to identify biomedical terms of interest (entities) and their relations. We also pretrained BERT on a corpus of 100,000 open access clinical publications and/or enhanced context-dependent entity classification with a conditional random field (CRF) model. Performance was measured using the F1 score, a metric that combines precision and recall. We defined successful matches as partial overlap of entities of the same type.
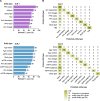
Results: For entity recognition, the pretrained BERT+CRF model had the best performance, with an F1 score of 73% in SLR 1 and 70% in SLR 2. Entity types identified with the highest accuracy were metrics for progression-free survival (SLR 1, F1 score 88%) or for patient age (SLR 2, F1 score 82%). Treatment arm dosage was identified less successfully (F1 scores 60% [SLR 1] and 49% [SLR 2]). The best-performing model for relation extraction, pretrained BERT relation classification, exhibited F1 scores higher than 90% in cases with at least 80 relation examples for a pair of related entity types.
Conclusions: The performance of BERT is enhanced by pretraining with biomedical literature and by combining with a CRF model. With refinement, machine learning may assist with manual data extraction for SLRs.
Keywords: Evidence-based practice; Information science; Information storage and retrieval; Methods; Systematic reviews as topic.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
All authors have completed the ICMJE uniform disclosure form at
Figures
Similar articles
-
Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records.BMC Med Inform Decis Mak. 2022 Mar 23;22(1):72. doi: 10.1186/s12911-022-01810-z. BMC Med Inform Decis Mak. 2022. PMID: 35321705 Free PMC article.
-
A Fine-Tuned Bidirectional Encoder Representations From Transformers Model for Food Named-Entity Recognition: Algorithm Development and Validation.J Med Internet Res. 2021 Aug 9;23(8):e28229. doi: 10.2196/28229. J Med Internet Res. 2021. PMID: 34383671 Free PMC article.
-
Extracting comprehensive clinical information for breast cancer using deep learning methods.Int J Med Inform. 2019 Dec;132:103985. doi: 10.1016/j.ijmedinf.2019.103985. Epub 2019 Oct 2. Int J Med Inform. 2019. PMID: 31627032
-
Machine learning models for abstract screening task - A systematic literature review application for health economics and outcome research.BMC Med Res Methodol. 2024 May 9;24(1):108. doi: 10.1186/s12874-024-02224-3. BMC Med Res Methodol. 2024. PMID: 38724903 Free PMC article.
-
Inter-reviewer reliability of human literature reviewing and implications for the introduction of machine-assisted systematic reviews: a mixed-methods review.BMJ Open. 2024 Mar 19;14(3):e076912. doi: 10.1136/bmjopen-2023-076912. BMJ Open. 2024. PMID: 38508610 Free PMC article. Review.
Cited by
-
The emergence of large language models as tools in literature reviews: a large language model-assisted systematic review.J Am Med Inform Assoc. 2025 Jun 1;32(6):1071-1086. doi: 10.1093/jamia/ocaf063. J Am Med Inform Assoc. 2025. PMID: 40332983 Free PMC article.
-
Testing the utility of GPT for title and abstract screening in environmental systematic evidence synthesis.Environ Evid. 2025 Apr 23;14(1):7. doi: 10.1186/s13750-025-00360-x. Environ Evid. 2025. PMID: 40270055 Free PMC article.
References
-
- National Institute for Health Research . PROSPERO: international prospective register of systematic reviews. 2011.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

