Analysis of chemical structures and mutations detected by Salmonella TA98 and TA100
- PMID: 37804576
- PMCID: PMC10841823
- DOI: 10.1016/j.mrfmmm.2023.111838
Analysis of chemical structures and mutations detected by Salmonella TA98 and TA100
Abstract
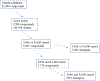
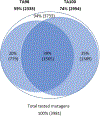
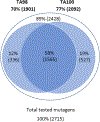
As part of an analysis performed under the auspices of the International Workshop on Genotoxicity Testing (IWGT) in 2017, we and others showed that Salmonella frameshift strain TA98 and base-substitution strain TA100 together + /- S9 detected 93% of the mutagens detected by all the bacterial strains recommended by OECD TG471 (Williams et al., Mutation Res. 848:503081, 2019). We have extended this analysis by identifying the numbers and chemical classes of chemicals detected by these two strains either alone or in combination, including the role of S9. Using the Leadscope 2021 SAR Genetox database containing > 21,900 compounds, our dataset containing 7170 compounds tested in both TA98 and TA100. Together, TA98 and TA100 detected 94% (3733/3981) of the mutagens detected using all the TG471-recommended bacterial strains; 39% were mutagenic in one or both strains. TA100 detected 77% of all of these mutagens and TA98 70%. Considering the overlap of detection by both strains, 12% of these mutagens were detected only by TA98 and 19% only by TA100. In the absence of S9, sensitivity dropped by 31% for TA98 and 29% for TA100. Overall, 32% of the mutagens required S9 for detection by either strain; 9% were detected only without S9. Using the 2021 Leadscope Genetox Expert Alerts, TA100 detected 18 mutagenic alerting chemical classes with better sensitivity than TA98, whereas TA98 detected 10 classes better than TA100. TA100 detected more chemical classes than did TA98, especially hydrazines, azides, various di- and tri-halides, various nitrosamines, epoxides, aziridines, difurans, and half-mustards; TA98 especially detected polycyclic primary amines, various aromatic amines, polycyclic aromatic hydrocarbons, triazines, and dibenzo-furans. Model compounds with these structures induce primarily G to T mutations in TA100 and/or a hotspot GC deletion in TA98. Both TA98 and TA100 + /- S9 are needed for adequate mutagenicity screening with the Salmonella (Ames) assay.
Keywords: Ames assay; Bacterial mutagenicity; Mutagenicity; Mutagenicity screening; Mutation spectra; Salmonella mutagenicity assay.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: David DeMarini has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Kevin P. Cross is employed by Instem and the databases used in this analysis were developed by and are the commercial property of Instem. The views expressed in this paper are those of the authors and do not reflect U.S. EPA policy.
Figures


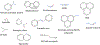

Similar articles
-
[Mutagenicity of mixtures of halogenated aliphatic hydrocarbons and polycyclic aromatic hydrocarbons in the Ames test with TA98 and TA100].Zentralbl Hyg Umweltmed. 1998 Feb;200(5-6):531-41. Zentralbl Hyg Umweltmed. 1998. PMID: 9531725 German.
-
Mutagens in surface waters: a review.Mutat Res. 2004 Nov;567(2-3):109-49. doi: 10.1016/j.mrrev.2004.08.003. Mutat Res. 2004. PMID: 15572284 Review.
-
A QSAR investigation of the role of hydrophobicity in regulating mutagenicity in the Ames test: 1. Mutagenicity of aromatic and heteroaromatic amines in Salmonella typhimurium TA98 and TA100.Environ Mol Mutagen. 1992;19(1):37-52. doi: 10.1002/em.2850190107. Environ Mol Mutagen. 1992. PMID: 1732103
-
Salmonella/human S9 mutagenicity test: a collaborative study with 58 compounds.Mutagenesis. 2005 May;20(3):217-28. doi: 10.1093/mutage/gei029. Epub 2005 Apr 20. Mutagenesis. 2005. PMID: 15843387
-
Revisiting the bacterial mutagenicity assays: Report by a workgroup of the International Workshops on Genotoxicity Testing (IWGT).Mutat Res Genet Toxicol Environ Mutagen. 2020 Jan;849:503137. doi: 10.1016/j.mrgentox.2020.503137. Epub 2020 Jan 13. Mutat Res Genet Toxicol Environ Mutagen. 2020. PMID: 32087853
Cited by
-
Synthesis and mutagenic risk of avanafil's potential genotoxic impurities.RSC Adv. 2024 Jul 8;14(30):21432-21438. doi: 10.1039/d4ra02345e. eCollection 2024 Jul 5. RSC Adv. 2024. PMID: 38979469 Free PMC article.
-
Mutagenic Atmospheres Generated from the Photooxidation of NOx with Selected VOCs and a Complex Mixture: Apportionment of Aromatic Mutagenicity for Reacted Gasoline Vapor.Atmos Environ (1994). 2024 Jun 25;334:120668. doi: 10.1016/j.atmosenv.2024.120668. Atmos Environ (1994). 2024. PMID: 39380947 Free PMC article.
-
Ames Assay Transferred from the Microtiter Plate to the Planar Assay Format.J Xenobiot. 2025 May 7;15(3):67. doi: 10.3390/jox15030067. J Xenobiot. 2025. PMID: 40407531 Free PMC article.
-
Molecular Compositions, Mutagenicity, and Mutation Spectra of Atmospheric Oxidation Products of Alkenes and Dienes Initiated by NOx + UV or Ozone: A Structure-Activity Analysis.Environ Sci Technol. 2024 Oct 22;58(42):18846-18855. doi: 10.1021/acs.est.4c04603. Epub 2024 Oct 7. Environ Sci Technol. 2024. PMID: 39374177
References
-
- Mortelmans K, Zeiger E, The Ames Salmonella/microsome mutagenicity assay, Mutation. Res 455 (2000) 29–60. - PubMed
-
- Patlewicz G, Dean JL, Gibbons CF, Judson RS, Keshava N, Vegosen L, Martin TM, Pradeep P, Simha A, Warren SH, Gwinn MR, DeMarini DM, Integrating publicly available information to screen potential candidates for chemical prioritization under the Toxic Substances Control Act: A proof of concept case study using genotoxicity and carcinogenicity, Comput. Toxicol 20 (2021) 1–100185. - PMC - PubMed
-
- REACH https://echa.europa.eu/regulations/reach/understanding-reach [Accessed 16 May 2023]
-
- OECD (2020). Test No. 471: Bacterial Reverse Mutation Test, OECD Guidelines for the Testing of Chemicals, Section 4, OECD Publishing, Paris, 10.787/9789264071247-en.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

