Ecological network analysis reveals cancer-dependent chaperone-client interaction structure and robustness
- PMID: 37805501
- PMCID: PMC10560210
- DOI: 10.1038/s41467-023-41906-2
Ecological network analysis reveals cancer-dependent chaperone-client interaction structure and robustness
Abstract
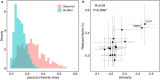
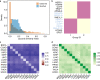
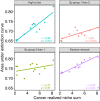
Cancer cells alter the expression levels of metabolic enzymes to fuel proliferation. The mitochondrion is a central hub of metabolic reprogramming, where chaperones service hundreds of clients, forming chaperone-client interaction networks. How network structure affects its robustness to chaperone targeting is key to developing cancer-specific drug therapy. However, few studies have assessed how structure and robustness vary across different cancer tissues. Here, using ecological network analysis, we reveal a non-random, hierarchical pattern whereby the cancer type modulates the chaperones' ability to realize their potential client interactions. Despite the low similarity between the chaperone-client interaction networks, we highly accurately predict links in one cancer type based on another. Moreover, we identify groups of chaperones that interact with similar clients. Simulations of network robustness show that this group structure affects cancer-specific response to chaperone removal. Our results open the door for new hypotheses regarding the ecology and evolution of chaperone-client interaction networks and can inform cancer-specific drug development strategies.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Voos W. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim. Biophys. Acta. 2013;1833:388–399. - PubMed
-
- Masgras I, et al. The molecular chaperone TRAP1 in cancer: from the basics of biology to pharmacological targeting. Semin. Cancer Biol. 2021;76:45–53. - PubMed
-
- Polson, E. S. et al. KHS101 disrupts energy metabolism in human glioblastoma cells and reduces tumor growth in mice. Sci. Transl. Med. 10, eaar2718 (2018). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

