Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10
- PMID: 37805510
- PMCID: PMC10560256
- DOI: 10.1038/s41467-023-42117-5
Open structure and gating of the Arabidopsis mechanosensitive ion channel MSL10
Abstract
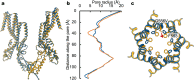
Plants are challenged by drastically different osmotic environments during growth and development. Adaptation to these environments often involves mechanosensitive ion channels that can detect and respond to mechanical force. In the model plant Arabidopsis thaliana, the mechanosensitive channel MSL10 plays a crucial role in hypo-osmotic shock adaptation and programmed cell death induction, but the molecular basis of channel function remains poorly understood. Here, we report a structural and electrophysiological analysis of MSL10. The cryo-electron microscopy structures reveal a distinct heptameric channel assembly. Structures of the wild-type channel in detergent and lipid environments, and in the absence of membrane tension, capture an open conformation. Furthermore, structural analysis of a non-conductive mutant channel demonstrates that reorientation of phenylalanine side chains alone, without main chain rearrangements, may generate the hydrophobic gate. Together, these results reveal a distinct gating mechanism and advance our understanding of mechanotransduction.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
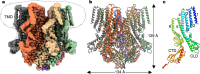



Similar articles
-
The Mechanosensitive Ion Channel MSL10 Potentiates Responses to Cell Swelling in Arabidopsis Seedlings.Curr Biol. 2020 Jul 20;30(14):2716-2728.e6. doi: 10.1016/j.cub.2020.05.015. Epub 2020 Jun 11. Curr Biol. 2020. PMID: 32531281
-
MscS-Like10 is a stretch-activated ion channel from Arabidopsis thaliana with a preference for anions.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):19015-20. doi: 10.1073/pnas.1213931109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112188 Free PMC article.
-
Cellular transduction of mechanical oscillations in plants by the plasma-membrane mechanosensitive channel MSL10.Proc Natl Acad Sci U S A. 2021 Jan 5;118(1):e1919402118. doi: 10.1073/pnas.1919402118. Proc Natl Acad Sci U S A. 2021. PMID: 33372153 Free PMC article.
-
Sensing and transducing forces in plants with MSL10 and DEK1 mechanosensors.FEBS Lett. 2018 Jun;592(12):1968-1979. doi: 10.1002/1873-3468.13102. Epub 2018 Jun 4. FEBS Lett. 2018. PMID: 29782638 Review.
-
From membrane tension to channel gating: A principal energy transfer mechanism for mechanosensitive channels.Protein Sci. 2016 Nov;25(11):1954-1964. doi: 10.1002/pro.3017. Epub 2016 Aug 23. Protein Sci. 2016. PMID: 27530280 Free PMC article. Review.
Cited by
-
A large-scale curated and filterable dataset for cryo-EM foundation model pre-training.Sci Data. 2025 Jun 7;12(1):960. doi: 10.1038/s41597-025-05179-2. Sci Data. 2025. PMID: 40483273 Free PMC article.
-
Lipid-mediated gating of a miniature mechanosensitive MscS channel from Trypanosoma cruzi.Nat Commun. 2025 Aug 8;16(1):7339. doi: 10.1038/s41467-025-62757-z. Nat Commun. 2025. PMID: 40781240 Free PMC article.
-
A mechanosensitive ion channel controls touch-triggered stigma movement through manipulation of calcium signature in Torenia.Nat Commun. 2025 Jul 8;16(1):6296. doi: 10.1038/s41467-025-61770-6. Nat Commun. 2025. PMID: 40628738 Free PMC article.
-
Lipid interactions and gating hysteresis suggest a physiological role for mechanosensitive channel YnaI.Nat Commun. 2025 Aug 12;16(1):7472. doi: 10.1038/s41467-025-62805-8. Nat Commun. 2025. PMID: 40796571 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

