Genomic architecture of autism spectrum disorder in Qatar: The BARAKA-Qatar Study
- PMID: 37805537
- PMCID: PMC10560429
- DOI: 10.1186/s13073-023-01228-w
Genomic architecture of autism spectrum disorder in Qatar: The BARAKA-Qatar Study
Abstract
Background: Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by impaired social and communication skills, restricted interests, and repetitive behaviors. The prevalence of ASD among children in Qatar was recently estimated to be 1.1%, though the genetic architecture underlying ASD both in Qatar and the greater Middle East has been largely unexplored. Here, we describe the first genomic data release from the BARAKA-Qatar Study-a nationwide program building a broadly consented biorepository of individuals with ASD and their families available for sample and data sharing and multi-omics research.
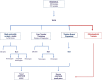
Methods: In this first release, we present a comprehensive analysis of whole-genome sequencing (WGS) data of the first 100 families (372 individuals), investigating the genetic architecture, including single-nucleotide variants (SNVs), copy number variants (CNVs), tandem repeat expansions (TREs), as well as mitochondrial DNA variants (mtDNA) segregating with ASD in local families.
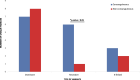
Results: Overall, we identify potentially pathogenic variants in known genes or regions in 27 out of 100 families (27%), of which 11 variants (40.7%) were classified as pathogenic or likely-pathogenic based on American College of Medical Genetics (ACMG) guidelines. Dominant variants, including de novo and inherited, contributed to 15 (55.6%) of these families, consisting of SNVs/indels (66.7%), CNVs (13.3%), TREs (13.3%), and mtDNA variants (6.7%). Moreover, homozygous variants were found in 7 families (25.9%), with a sixfold increase in homozygous burden in consanguineous versus non-consanguineous families (13.6% and 1.8%, respectively). Furthermore, 28 novel ASD candidate genes were identified in 20 families, 23 of which had recurrent hits in MSSNG and SSC cohorts.
Conclusions: This study illustrates the value of ASD studies in under-represented populations and the importance of WGS as a comprehensive tool for establishing a molecular diagnosis for families with ASD. Moreover, it uncovers a significant role for recessive variation in ASD architecture in consanguineous settings and provides a unique resource of Middle Eastern genomes for future research to the global ASD community.
Keywords: ASD; ASD risk genes; Autism spectrum disorder; BARAKA cohort; De novo variants; Middle Eastern population; SNVs; Whole genome sequencing.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
S.W.S. is a scientific advisor to Population Bio and a Highly Cited Academic Advisor to the King Abdullaziz University. Intellectual property from his work held at the University of Toronto has been licensed to Athena Diagnostics. None of these relationships have influence the research described in this paper. The remaining authors declare that they have no competing interests.
Figures


References
-
- American Psychiatric Association. DSM-5 Autism Spectrum Disorder Fact Sheet. Am Psychiatr Assoc. 2014;(October):233–55.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

