URI alleviates tyrosine kinase inhibitors-induced ferroptosis by reprogramming lipid metabolism in p53 wild-type liver cancers
- PMID: 37805657
- PMCID: PMC10560259
- DOI: 10.1038/s41467-023-41852-z
URI alleviates tyrosine kinase inhibitors-induced ferroptosis by reprogramming lipid metabolism in p53 wild-type liver cancers
Abstract
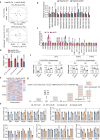
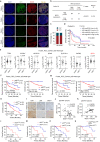
The clinical benefit of tyrosine kinase inhibitors (TKIs)-based systemic therapy for advanced hepatocellular carcinoma (HCC) is limited due to drug resistance. Here, we uncover that lipid metabolism reprogramming mediated by unconventional prefoldin RPB5 interactor (URI) endows HCC with resistance to TKIs-induced ferroptosis. Mechanistically, URI directly interacts with TRIM28 and promotes p53 ubiquitination and degradation in a TRIM28-MDM2 dependent manner. Importantly, p53 binds to the promoter of stearoyl-CoA desaturase 1 (SCD1) and represses its transcription. High expression of URI is correlated with high level of SCD1 and their synergetic expression predicts poor prognosis and TKIs resistance in HCC. The combination of SCD1 inhibitor aramchol and deuterated sorafenib derivative donafenib displays promising anti-tumor effects in p53-wild type HCC patient-derived organoids and xenografted tumors. This combination therapy has potential clinical benefits for the patients with advanced HCC who have wild-type p53 and high levels of URI/SCD1.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declared no competing interests.
Figures







References
-
- Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–1362. - PubMed
-
- Llovet JM, et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. - PubMed
-
- Bruix J, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

