Side-chain dynamics of the α1B -adrenergic receptor determined by NMR via methyl relaxation
- PMID: 37805830
- PMCID: PMC10593183
- DOI: 10.1002/pro.4801
Side-chain dynamics of the α1B -adrenergic receptor determined by NMR via methyl relaxation
Abstract
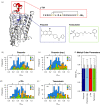
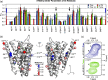
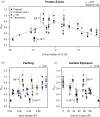
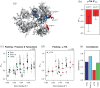
G protein-coupled receptors (GPCRs) are medically important membrane proteins that sample inactive, intermediate, and active conformational states characterized by relatively slow interconversions (~μs-ms). On a faster timescale (~ps-ns), the conformational landscape of GPCRs is governed by the rapid dynamics of amino acid side chains. Such dynamics are essential for protein functions such as ligand recognition and allostery. Unfortunately, technical challenges have almost entirely precluded the study of side-chain dynamics for GPCRs. Here, we investigate the rapid side-chain dynamics of a thermostabilized α1B -adrenergic receptor (α1B -AR) as probed by methyl relaxation. We determined order parameters for Ile, Leu, and Val methyl groups in the presence of inverse agonists that bind orthosterically (prazosin, tamsulosin) or allosterically (conopeptide ρ-TIA). Despite the differences in the ligands, the receptor's overall side-chain dynamics are very similar, including those of the apo form. However, ρ-TIA increases the flexibility of Ile1764×56 and possibly of Ile2145×49 , adjacent to Pro2155×50 of the highly conserved P5×50 I3×40 F6×44 motif crucial for receptor activation, suggesting differences in the mechanisms for orthosteric and allosteric receptor inactivation. Overall, increased Ile side-chain rigidity was found for residues closer to the center of the membrane bilayer, correlating with denser packing and lower protein surface exposure. In contrast to two microbial membrane proteins, in α1B -AR Leu exhibited higher flexibility than Ile side chains on average, correlating with the presence of Leu in less densely packed areas and with higher protein-surface exposure than Ile. Our findings demonstrate the feasibility of studying receptor-wide side-chain dynamics in GPCRs to gain functional insights.
Keywords: GPCR; allosteric ligand; inverse agonist; membrane protein; order parameter.
© 2023 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Baumann C, Zerbe O. The role of leucine and isoleucine in tuning the hydropathy of class A GPCRs. Proteins. 2023. - PubMed
-
- Best RB, Clarke J, Karplus M. The origin of protein sidechain order parameter distributions. J Am Chem Soc. 2004;126(25):7734–7735. - PubMed
-
- Chen X, Xu Y, Qu L, Wu L, Han G, Guo Y, et al. Molecular mechanism for ligand recognition and subtype selectivity of α2C adrenergic receptor. Cell Rep. 2019;29(10):2936–2943.e2934. - PubMed
-
- Chen Z, Rogge G, Hague C, Alewood D, Colless B, Lewis RJ, et al. Subtype‐selective noncompetitive or competitive inhibition of human α1‐adrenergic receptors by ρ‐TIA. J Biol Chem. 2004;279(34):35326–35333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

