Free and Interfacial Boundaries in Individual-Based Models of Multicellular Biological systems
- PMID: 37805982
- PMCID: PMC10560655
- DOI: 10.1007/s11538-023-01214-8
Free and Interfacial Boundaries in Individual-Based Models of Multicellular Biological systems
Abstract
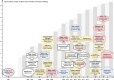
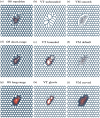
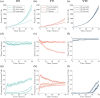
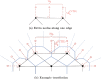
Coordination of cell behaviour is key to a myriad of biological processes including tissue morphogenesis, wound healing, and tumour growth. As such, individual-based computational models, which explicitly describe inter-cellular interactions, are commonly used to model collective cell dynamics. However, when using individual-based models, it is unclear how descriptions of cell boundaries affect overall population dynamics. In order to investigate this we define three cell boundary descriptions of varying complexities for each of three widely used off-lattice individual-based models: overlapping spheres, Voronoi tessellation, and vertex models. We apply our models to multiple biological scenarios to investigate how cell boundary description can influence tissue-scale behaviour. We find that the Voronoi tessellation model is most sensitive to changes in the cell boundary description with basic models being inappropriate in many cases. The timescale of tissue evolution when using an overlapping spheres model is coupled to the boundary description. The vertex model is demonstrated to be the most stable to changes in boundary description, though still exhibits timescale sensitivity. When using individual-based computational models one should carefully consider how cell boundaries are defined. To inform future work, we provide an exploration of common individual-based models and cell boundary descriptions in frequently studied biological scenarios and discuss their benefits and disadvantages.
Keywords: Cell boundaries; Individual-based models; Tissue growth; Wound healing.
© 2023. The Author(s).
Figures









References
-
- Bock M, Tyagi AK, Kreft J-U, Alt W. Generalized voronoi tessellation as a model of two-dimensional cell tissue dynamics. Bull Math Biol. 2010;72(7):1696–1731. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

