Viruses of the Turriviridae: an emerging model system for studying archaeal virus-host interactions
- PMID: 37808280
- PMCID: PMC10551542
- DOI: 10.3389/fmicb.2023.1258997
Viruses of the Turriviridae: an emerging model system for studying archaeal virus-host interactions
Abstract
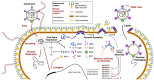
Viruses have played a central role in the evolution and ecology of cellular life since it first arose. Investigations into viral molecular biology and ecological dynamics have propelled abundant progress in our understanding of living systems, including genetic inheritance, cellular signaling and trafficking, and organismal development. As well, the discovery of viral lineages that infect members of all three domains suggest that these lineages originated at the earliest stages of biological evolution. Research into these viruses is helping to elucidate the conditions under which life arose, and the dynamics that directed its early development. Archaeal viruses have only recently become a subject of intense study, but investigations have already produced intriguing and exciting results. STIV was originally discovered in Yellowstone National Park and has been the focus of concentrated research. Through this research, a viral genetic system was created, a novel lysis mechanism was discovered, and the interaction of the virus with cellular ESCRT machinery was revealed. This review will summarize the discoveries within this group of viruses and will also discuss future work.
Keywords: Sulfolobus turreted icosahedral virus; Turriviridae; archaea; viral replication; virus; virus-host interaction.
Copyright © 2023 Overton, Manuel, Lawrence and Snyder.
Conflict of interest statement
The authors declare that the research performed for writing this review was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

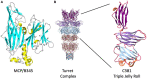

References
-
- Albers S. V., Siebers B. (2014). “The family Sulfolobaceae” in The prokaryotes. eds. Rosenberg E., et al.. 4th ed (Berlin, Heidelberg: Springer-Verlag; ), 323–346. doi: 10.1007/978-3-642-38954-2_329 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

