Machine learning approaches in microbiome research: challenges and best practices
- PMID: 37808286
- PMCID: PMC10556866
- DOI: 10.3389/fmicb.2023.1261889
Machine learning approaches in microbiome research: challenges and best practices
Abstract
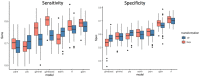
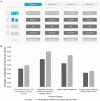
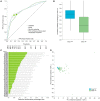
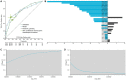
Microbiome data predictive analysis within a machine learning (ML) workflow presents numerous domain-specific challenges involving preprocessing, feature selection, predictive modeling, performance estimation, model interpretation, and the extraction of biological information from the results. To assist decision-making, we offer a set of recommendations on algorithm selection, pipeline creation and evaluation, stemming from the COST Action ML4Microbiome. We compared the suggested approaches on a multi-cohort shotgun metagenomics dataset of colorectal cancer patients, focusing on their performance in disease diagnosis and biomarker discovery. It is demonstrated that the use of compositional transformations and filtering methods as part of data preprocessing does not always improve the predictive performance of a model. In contrast, the multivariate feature selection, such as the Statistically Equivalent Signatures algorithm, was effective in reducing the classification error. When validated on a separate test dataset, this algorithm in combination with random forest modeling, provided the most accurate performance estimates. Lastly, we showed how linear modeling by logistic regression coupled with visualization techniques such as Individual Conditional Expectation (ICE) plots can yield interpretable results and offer biological insights. These findings are significant for clinicians and non-experts alike in translational applications.
Keywords: AutoML; colorectal cancer; feature selection; machine learning methods; microbiome data analysis; model selection; predictive modeling; preprocessing.
Copyright © 2023 Papoutsoglou, Tarazona, Lopes, Klammsteiner, Ibrahimi, Eckenberger, Novielli, Tonda, Simeon, Shigdel, Béreux, Vitali, Tangaro, Lahti, Temko, Claesson and Berland.
Conflict of interest statement
GP was directly affiliated with JADBio—Gnosis DA, S.A., which offers the JADBio service commercially. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Aitchison J. (1982). The statistical analysis of compositional data. J. R. Stat. Soc. B 44, 139–160. doi: 10.1111/j.2517-6161.1982.tb01195.x - DOI
-
- Akosa J. (2017). Predictive accuracy: a misleading performance measure for highly imbalanced data. Availanble at: https://www.semanticscholar.org/paper/Predictive-Accuracy-%3A-A-Misleadi...
-
- Barbet P., Almeida M., Probul N., Baumach J., Pons N., Plaza Onate F., et al. (2023). Taxonomic profiles, functional profiles and manually curated metadata of human fecal metagenomes from public projects coming from colorectal cancer studies (version 5) [dataset]. Recher. Data Gouv. doi: 10.57745/7IVO3E - DOI
Publication types
LinkOut - more resources
Full Text Sources

