Genomic epidemiology and characterization of Staphylococcus aureus isolates from raw milk in Jiangsu, China: emerging broader host tropism strain clones ST59 and ST398
- PMID: 37808296
- PMCID: PMC10556526
- DOI: 10.3389/fmicb.2023.1266715
Genomic epidemiology and characterization of Staphylococcus aureus isolates from raw milk in Jiangsu, China: emerging broader host tropism strain clones ST59 and ST398
Abstract
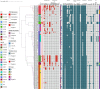
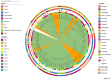
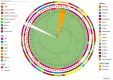
Staphylococcus aureus is highly pathogenic and can cause disease in both humans and domestic animals. The aim of this study was to investigate the genomic epidemiology of S. aureus isolates from raw milk in Jiangsu Province, China, to identify predominant lineages and their associated genomic and phenotypic characteristics. In this study, we identified 117 S. aureus isolates collected from 1,062 samples in Jiangsu Province between 2021 and 2022. Based on whole-genome sequencing (WGS) data from 117 S. aureus isolates, molecular analyses indicated CC1-ST1 (26.50%, 31/117), CC97-ST97 (18.80%, 22/117), CC398-ST398 (10.26%, 12/117), CC8-ST630 (7.69%, 9/117) and CC59-ST59 (2.56%, 3/117) were the major lineages. The prevalence of mecA-positive strains was 11.11%. Four methicillin-resistant S. aureus (MRSA) lineages were found, including MRSA-ST59-t172 (n = 3), OS-MRSA-ST398-t011 (n = 1), MRSA-ST630-t2196 (n = 2) and OS-MRSA-ST630-t2196 (n = 7). Phenotypic resistance to penicillin (30.77%, 36/117), ciprofloxacin (17.09%, 20/117) and erythromycin (15.38%, 18/117) was observed which corresponded with resistance genotypes. All of the isolates could produce biofilms, and 38.46% (45/117) of isolates had invasion rates in mammary epithelial cells (MAC-T) of greater than 1%. Interestingly, most biofilm-producing and invading isolates harbored ebp-icaA-icaB-icaC-icaR-clfA-clfB-fnbA-fnbB-sdrC-sdrD-sdrE-map-can (27.35%, 32/117) and ebp-icaA-icaB-icaC-icaD-icaR-clfA-clfB-fnbA-fnbB-sdrC-sdrD-sdrE-map (33.33%, 39/117) adherence-associated gene patterns and belonged to lineages CC1 and CC97, respectively. Virulence factor assays showed that 47.01% of the isolates contained at least enterotoxin genes. Isolates harboring the immune evasion cluster (IEC) genes (sea, sak, chp, and scn) were predominantly categorized as STs 464, 398, and 59. IEC-positive ST398 and ST59 isolates contained a very high proportion of virulence genes located on prophages, whereas most IEC-negative ST398 clade isolates carried broad-spectrum drug resistance genes. Meanwhile, the IEC-positive ST398 clade showed a close genetic relationship with isolates from the pork supply chain and hospital surgical site infections. MRSA-ST59 strains showed the closest genetic relationship with an isolate from quick-frozen products. High-risk livestock-associated strains ST398 and MRSA-ST59 were detected in raw milk, indicating a potential public health risk of S. aureus transmission between livestock and humans. Our study highlights the necessity for S. aureus surveillance in the dairy industry.
Keywords: Staphylococcus aureus; biofilm formation; invasion; phylogenetic analysis; virulence factors; whole-genome sequencing.
Copyright © 2023 Liu, Ji, Wang, Hou, Sun, Billington, Zhang, Wang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ahmadrajabi R., Layegh-Khavidaki S., Kalantar-Neyestanaki D., Fasihi Y. (2017). Molecular analysis of immune evasion cluster (IEC) genes and intercellular adhesion gene cluster (ICA) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J. Prev. Med. Hyg. 58, E308–e314. doi: 10.15167/2421-4248/jpmh2017.58.4.711, PMID: - DOI - PMC - PubMed
-
- Algammal A. M., Enany M. E., El-Tarabili R. M., Ghobashy M. O. I., Helmy Y. A. (2020). Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens 9:362. doi: 10.3390/pathogens9050362, PMID: - DOI - PMC - PubMed
-
- Bazari P. A. M., Honarmand Jahromy S., Zare Karizi S. (2017). Phenotypic and genotypic characterization of biofilm formation among Staphylococcus aureus isolates from clinical specimens, an atomic force microscopic (AFM) study. Microb. Pathog. 110, 533–539. doi: 10.1016/j.micpath.2017.07.041 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

