Hepatitis C virus alters the morphology and function of peroxisomes
- PMID: 37808318
- PMCID: PMC10551450
- DOI: 10.3389/fmicb.2023.1254728
Hepatitis C virus alters the morphology and function of peroxisomes
Abstract
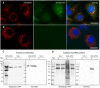
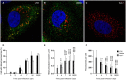
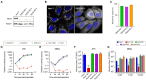
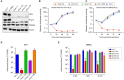
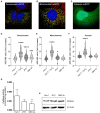
Despite the introduction of effective treatments for hepatitis C in clinics, issues remain regarding the liver disease induced by chronic hepatitis C virus (HCV) infection. HCV is known to disturb the metabolism of infected cells, especially lipid metabolism and redox balance, but the mechanisms leading to HCV-induced pathogenesis are still poorly understood. In an APEX2-based proximity biotinylation screen, we identified ACBD5, a peroxisome membrane protein, as located in the vicinity of HCV replication complexes. Confocal microscopy confirmed the relocation of peroxisomes near HCV replication complexes and indicated that their morphology and number are altered in approximately 30% of infected Huh-7 cells. Peroxisomes are small versatile organelles involved among other functions in lipid metabolism and ROS regulation. To determine their importance in the HCV life cycle, we generated Huh-7 cells devoid of peroxisomes by inactivating the PEX5 and PEX3 genes using CRISPR/Cas9 and found that the absence of peroxisomes had no impact on replication kinetics or infectious titers of HCV strains JFH1 and DBN3a. The impact of HCV on peroxisomal functions was assessed using sub-genomic replicons. An increase of ROS was measured in peroxisomes of replicon-containing cells, correlated with a significant decrease of catalase activity with the DBN3a strain. In contrast, HCV replication had little to no impact on cytoplasmic and mitochondrial ROS, suggesting that the redox balance of peroxisomes is specifically impaired in cells replicating HCV. Our study provides evidence that peroxisome function and morphology are altered in HCV-infected cells.
Keywords: APEX2; CRISPR-Cas9; HCV genotype; ROS; hepatitis C virus; peroxisome; proximity biotinylation.
Copyright © 2023 Martin de Fourchambault, Callens, Saliou, Fourcot, Delos, Barois, Thorel, Ramirez, Bukh, Cocquerel, Bertrand-Michel, Marot, Sebti, Dubuisson and Rouillé.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ali R. O., Quinn G. M., Umarova R., Haddad J. A., Zhang G. Y., Townsend E. C., et al. (2023). Longitudinal multi-omics analyses of the gut-liver axis reveals metabolic dysregulation in hepatitis C infection and cirrhosis. Nat. Microbiol. 8, 12–27. doi: 10.1038/s41564-022-01273-y, PMID: - DOI - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials

