This is a preprint.
A suite of engineered mice for interrogating psychedelic drug actions
- PMID: 37808655
- PMCID: PMC10557740
- DOI: 10.1101/2023.09.25.559347
A suite of engineered mice for interrogating psychedelic drug actions
Abstract
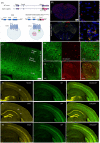
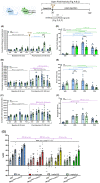
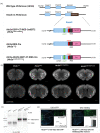
Psychedelic drugs like lysergic acid diethylamide (LSD) and psilocybin have emerged as potentially transformative therapeutics for many neuropsychiatric diseases, including depression, anxiety, post-traumatic stress disorder, migraine, and cluster headaches. LSD and psilocybin exert their psychedelic effects via activation of the 5-hydroxytryptamine 2A receptor (HTR2A). Here we provide a suite of engineered mice useful for clarifying the role of HTR2A and HTR2A-expressing neurons in psychedelic drug actions. We first generated Htr2a-EGFP-CT-IRES-CreERT2 mice (CT:C-terminus) to independently identify both HTR2A-EGFP-CT receptors and HTR2A-containing cells thereby providing a detailed anatomical map of HTR2A and identifying cell types that express HTR2A. We also generated a humanized Htr2a mouse line and an additional constitutive Htr2A-Cre mouse line. Psychedelics induced a variety of known behavioral changes in our mice validating their utility for behavioral studies. Finally, electrophysiology studies revealed that extracellular 5-HT elicited a HTR2A-mediated robust increase in firing of genetically-identified pyramidal neurons--consistent with a plasma membrane localization and mode of action. These mouse lines represent invaluable tools for elucidating the molecular, cellular, pharmacological, physiological, behavioral, and other actions of psychedelic drugs in vivo.
Figures




Similar articles
-
Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.Neuron. 2022 Oct 5;110(19):3154-3167.e7. doi: 10.1016/j.neuron.2022.08.006. Epub 2022 Sep 9. Neuron. 2022. PMID: 36087581 Free PMC article.
-
New Paradigms of Old Psychedelics in Schizophrenia.Pharmaceuticals (Basel). 2022 May 23;15(5):640. doi: 10.3390/ph15050640. Pharmaceuticals (Basel). 2022. PMID: 35631466 Free PMC article. Review.
-
Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor.Cell. 2020 Sep 17;182(6):1574-1588.e19. doi: 10.1016/j.cell.2020.08.024. Cell. 2020. PMID: 32946782 Free PMC article.
-
Psychedelics: Alternative and Potential Therapeutic Options for Treating Mood and Anxiety Disorders.Molecules. 2022 Apr 14;27(8):2520. doi: 10.3390/molecules27082520. Molecules. 2022. PMID: 35458717 Free PMC article. Review.
-
Htr2a Gene and 5-HT(2A) Receptor Expression in the Cerebral Cortex Studied Using Genetically Modified Mice.Front Neurosci. 2010 Aug 13;4:36. doi: 10.3389/fnins.2010.00036. eCollection 2010. Front Neurosci. 2010. PMID: 20802802 Free PMC article.
References
-
- Backstrom J.R., and Sanders-Bush E. (1997). Generation of anti-peptide antibodies against serotonin 5HT2A and 5-HT2C receptors. J Neurosci Methods 77, 109–117. - PubMed
-
- Barnes N.M., Ahern G.P., Becamel C., Bockaert J., Camilleri M., Chaumont-Dubel S., Claeysen S., Cunningham K.A., Fone K.C., Gershon M., Di Giovanni G., Goodfellow N.M., Halberstadt A.L., Hartley R.M., Hassaine G., Herrick-Davis K., Hovius R., Lacivita E., Lambe E.K., Leopoldo M., Levy F.O., Lummis S.C.R., Marin P., Maroteaux L., McCreary A.C., Nelson D.L., Neumaier J.F., Newman-Tancredi A., Nury H., Roberts A., Roth B.L., Roumier A., Sanger G.J., Teitler M., Sharp T., Villalon C.M., Vogel H., Watts S.W., and Hoyer D. (2021). International Union of Basic and Clinical Pharmacology. CX. Classification of Receptors for 5-hydroxytryptamine; Pharmacology and Function. Pharmacol Rev 73, 310–520. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
