This is a preprint.
Germline mutations and developmental mosaicism underlying EGFR-mutant lung cancer
- PMID: 37808694
- PMCID: PMC10557804
- DOI: 10.1101/2023.09.28.23296274
Germline mutations and developmental mosaicism underlying EGFR-mutant lung cancer
Update in
-
Developmental mosaicism underlying EGFR-mutant lung cancer presenting with multiple primary tumors.Nat Cancer. 2024 Nov;5(11):1681-1696. doi: 10.1038/s43018-024-00840-y. Epub 2024 Oct 15. Nat Cancer. 2024. PMID: 39406916 Free PMC article.
Abstract
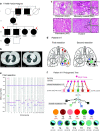
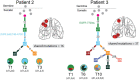
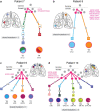
While the development of multiple primary tumors in smokers with lung cancer can be attributed to carcinogen-induced field cancerization, the occurrence of multiple primary tumors in individuals with EGFR-mutant lung cancer who lack known environmental exposures remains unexplained. We identified ten patients with early-stage, resectable non-small cell lung cancer who presented with multiple anatomically distinct EGFR-mutant tumors. We analyzed the phylogenetic relationships among multiple tumors from each patient using whole exome sequencing (WES) and hypermutable poly-guanine (poly-G) repeat genotyping, as orthogonal methods for lineage tracing. In two patients, we identified germline EGFR variants, which confer moderately enhanced signaling when modeled in vitro. In four other patients, developmental mosaicism is supported by the poly-G lineage tracing and WES, indicating a common non-germline cell-of-origin. Thus, developmental mosaicism and germline variants define two distinct mechanisms of genetic predisposition to multiple EGFR-mutant primary tumors, with implications for understanding their etiology and clinical management.
Figures



References
-
- Detterbeck F.C., et al. The IASLC Lung Cancer Staging Project: Summary of Proposals for Revisions of the Classification of Lung Cancers with Multiple Pulmonary Sites of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 11, 639–650 (2016). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
