This is a preprint.
Prognostic Insights from Longitudinal Multicompartment Study of Host-Microbiota Interactions in Critically Ill Patients
- PMID: 37808745
- PMCID: PMC10557814
- DOI: 10.1101/2023.09.25.23296086
Prognostic Insights from Longitudinal Multicompartment Study of Host-Microbiota Interactions in Critically Ill Patients
Abstract
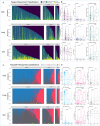
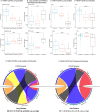
Critical illness can disrupt the composition and function of the microbiome, yet comprehensive longitudinal studies are lacking. We conducted a longitudinal analysis of oral, lung, and gut microbiota in a large cohort of 479 mechanically ventilated patients with acute respiratory failure. Progressive dysbiosis emerged in all three body compartments, characterized by reduced alpha diversity, depletion of obligate anaerobe bacteria, and pathogen enrichment. Clinical variables, including chronic obstructive pulmonary disease, immunosuppression, and antibiotic exposure, shaped dysbiosis. Notably, of the three body compartments, unsupervised clusters of lung microbiota diversity and composition independently predicted survival, transcending clinical predictors, organ dysfunction severity, and host-response sub-phenotypes. These independent associations of lung microbiota may serve as valuable biomarkers for prognostication and treatment decisions in critically ill patients. Insights into the dynamics of the microbiome during critical illness highlight the potential for microbiota-targeted interventions in precision medicine.
Keywords: biomarkers; critical illness; dysbiosis; microbiome; precision medicine.
Conflict of interest statement
Conflicts of Interest: Dr. Kitsios has received research funding from Karius, Inc and Pfizer, Inc, both unrelated to this project. Dr. Morris has received research funding from Pfizer, Inc, unrelated to this project. Dr McVerry has received consulting fees from Boehringer Ingelheim, BioAegis, and Synairgen Research, Ltd. unrelated to this work. All other authors disclosed no conflict of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
