This is a preprint.
Electrostatic Interactions Dictate Bile Salt Hydrolase Substrate Preference
- PMID: 37808785
- PMCID: PMC10557579
- DOI: 10.1101/2023.09.25.559308
Electrostatic Interactions Dictate Bile Salt Hydrolase Substrate Preference
Update in
-
Electrostatic Interactions Dictate Bile Salt Hydrolase Substrate Preference.Biochemistry. 2023 Nov 7;62(21):3076-3084. doi: 10.1021/acs.biochem.3c00210. Epub 2023 Oct 26. Biochemistry. 2023. PMID: 37883888 Free PMC article.
Abstract
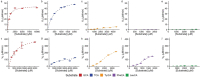
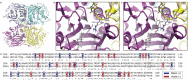
The human intestines are colonized by trillions of microbes, comprising the gut microbiota, which produce diverse small molecule metabolites and modify host metabolites, such as bile acids, that regulate host physiology. Biosynthesized in the liver, bile acids are conjugated with glycine or taurine and secreted into the intestines, where gut microbial bile salt hydrolases (BSHs) deconjugate the amino acid to produce unconjugated bile acids that serve as precursors for secondary bile acid metabolites. Among these include a recently discovered class of microbially-conjugated bile acids (MCBAs), wherein alternative amino acids are conjugated onto bile acids. To elucidate the metabolic potential of MCBAs, we performed detailed kinetic studies to investigate the preference of BSHs for host- and microbially-conjugated bile acids. We identified a BSH that exhibits positive cooperativity uniquely for MCBAs containing an aromatic sidechain. Further molecular modeling and phylogenetic analyses indicated that BSH preference for aromatic MCBAs is due to a substrate-specific cation-π interaction and is predicted to be widespread among human gut microbial BSHs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Electrostatic Interactions Dictate Bile Salt Hydrolase Substrate Preference.Biochemistry. 2023 Nov 7;62(21):3076-3084. doi: 10.1021/acs.biochem.3c00210. Epub 2023 Oct 26. Biochemistry. 2023. PMID: 37883888 Free PMC article.
-
Bile salt hydrolase acyltransferase activity expands bile acid diversity.Nature. 2024 Feb;626(8000):852-858. doi: 10.1038/s41586-024-07017-8. Epub 2024 Feb 7. Nature. 2024. PMID: 38326608
-
Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.Nat Microbiol. 2023 Apr;8(4):611-628. doi: 10.1038/s41564-023-01337-7. Epub 2023 Mar 13. Nat Microbiol. 2023. PMID: 36914755 Free PMC article.
-
New insights into microbial bile salt hydrolases: from physiological roles to potential applications.Front Microbiol. 2025 Feb 12;16:1513541. doi: 10.3389/fmicb.2025.1513541. eCollection 2025. Front Microbiol. 2025. PMID: 40012771 Free PMC article. Review.
-
Targeting gut microbial bile salt hydrolase (BSH) by diet supplements: new insights into dietary modulation of human health.Food Funct. 2022 Jul 18;13(14):7409-7422. doi: 10.1039/d2fo01252a. Food Funct. 2022. PMID: 35766281 Review.
References
-
- Ridlon J. M.; Kang D.-J.; Hylemon P. B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47 (2), 241–259. - PubMed
-
- Quinn R. A.; Melnik A. V.; Vrbanac A.; Fu T.; Patras K. A.; Christy M. P.; Bodai Z.; Belda-Ferre P.; Tripathi A.; Chung L. K.; Downes M.; Welch R. D.; Quinn M.; Humphrey G.; Panitchpakdi M.; Weldon K. C.; Aksenov A.; da Silva R.; Avila-Pacheco J.; Clish C.; Bae S.; Mallick H.; Franzosa E. A.; Lloyd-Price J.; Bussell R.; Thron T.; Nelson A. T.; Wang M.; Leszczynski E.; Vargas F.; Gauglitz J. M.; Meehan M. J.; Gentry E.; Arthur T. D.; Komor A. C.; Poulsen O.; Boland B. S.; Chang J. T.; Sandborn W. J.; Lim M.; Garg N.; Lumeng J. C.; Xavier R. J.; Kazmierczak B. I.; Jain R.; Egan M.; Rhee K. E.; Ferguson D.; Raffatellu M.; Vlamakis H.; Haddad G. G.; Siegel D.; Huttenhower C.; Mazmanian S. K.; Evans R. M.; Nizet V.; Knight R.; Dorrestein P. C. Global Chemical Effects of the Microbiome Include New Bile-Acid Conjugations. Nature 2020, 579 (7797), 123–129. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
