PheWAS and cross-disorder analysis reveal genetic architecture, pleiotropic loci and phenotypic correlations across 11 autoimmune disorders
- PMID: 37809097
- PMCID: PMC10552152
- DOI: 10.3389/fimmu.2023.1147573
PheWAS and cross-disorder analysis reveal genetic architecture, pleiotropic loci and phenotypic correlations across 11 autoimmune disorders
Abstract
Introduction: Autoimmune disorders (ADs) are a group of about 80 disorders that occur when self-attacking autoantibodies are produced due to failure in the self-tolerance mechanisms. ADs are polygenic disorders and associations with genes both in the human leukocyte antigen (HLA) region and outside of it have been described. Previous studies have shown that they are highly comorbid with shared genetic risk factors, while epidemiological studies revealed associations between various lifestyle and health-related phenotypes and ADs.
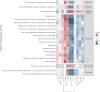
Methods: Here, for the first time, we performed a comparative polygenic risk score (PRS) - Phenome Wide Association Study (PheWAS) for 11 different ADs (Juvenile Idiopathic Arthritis, Primary Sclerosing Cholangitis, Celiac Disease, Multiple Sclerosis, Rheumatoid Arthritis, Psoriasis, Myasthenia Gravis, Type 1 Diabetes, Systemic Lupus Erythematosus, Vitiligo Late Onset, Vitiligo Early Onset) and 3,254 phenotypes available in the UK Biobank that include a wide range of socio-demographic, lifestyle and health-related outcomes. Additionally, we investigated the genetic relationships of the studied ADs, calculating their genetic correlation and conducting cross-disorder GWAS meta-analyses for the observed AD clusters.
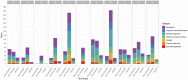
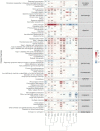
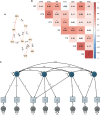
Results: In total, we identified 508 phenotypes significantly associated with at least one AD PRS. 272 phenotypes were significantly associated after excluding variants in the HLA region from the PRS estimation. Through genetic correlation and genetic factor analyses, we identified four genetic factors that run across studied ADs. Cross-trait meta-analyses within each factor revealed pleiotropic genome-wide significant loci.
Discussion: Overall, our study confirms the association of different factors with genetic susceptibility for ADs and reveals novel observations that need to be further explored.
Keywords: GWAS; PRS; PheWAS; autoimmune disorders; cross-disorder; meta-analysis.
Copyright © 2023 Topaloudi, Jain, Martinez, Bryant, Reynolds, Zagoriti, Lagoumintzis, Zamba-Papanicolaou, Tzartos, Poulas, Kleopa, Tzartos, Georgitsi, Drineas and Paschou.
Conflict of interest statement
KP is Founder and Head of the Institute for Research and Innovation. Based at Patras Science Park. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

