Occurrence of SARS-CoV-2 viremia is associated with genetic variants of genes related to COVID-19 pathogenesis
Affiliations
- 1 Internal Medicine Department, Hospital Universitario La Princesa, Madrid, Spain.
- 2 Instituto de Investigación Sanitaria La Princesa (IIS-IP), Madrid, Spain.
- 3 Rheumathology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 4 Molecular Biology Unit, Hospital Universitario La Princesa, Madrid, Spain.
- 5 Microbiology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 6 Immunology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 7 Cell Biology Department, Facultad de Biología, Universidad Complutense, Madrid, Spain.
- 8 Clinical Pharmacology Department, Hospital Universitario La Princesa, Instituto Teófilo Hernando, Universidad Autónoma de Madrid (UAM), Madrid, Spain.
- 9 Pneumology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 10 Anesthesiology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 11 Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, Spain.
- 12 Centro de Investigación Biomédica en Red en Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, Spain.
- 13 Intensive Care Unit, Hospital Universitario La Princesa, Madrid, Spain.
- 14 Universidad Autónoma de Madrid (UAM), Madrid, Spain.
- PMID: 37809329
- PMCID: PMC10557488
- DOI: 10.3389/fmed.2023.1215246
Occurrence of SARS-CoV-2 viremia is associated with genetic variants of genes related to COVID-19 pathogenesis
Authors
Affiliations
- 1 Internal Medicine Department, Hospital Universitario La Princesa, Madrid, Spain.
- 2 Instituto de Investigación Sanitaria La Princesa (IIS-IP), Madrid, Spain.
- 3 Rheumathology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 4 Molecular Biology Unit, Hospital Universitario La Princesa, Madrid, Spain.
- 5 Microbiology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 6 Immunology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 7 Cell Biology Department, Facultad de Biología, Universidad Complutense, Madrid, Spain.
- 8 Clinical Pharmacology Department, Hospital Universitario La Princesa, Instituto Teófilo Hernando, Universidad Autónoma de Madrid (UAM), Madrid, Spain.
- 9 Pneumology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 10 Anesthesiology Department, Hospital Universitario La Princesa, Madrid, Spain.
- 11 Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, Spain.
- 12 Centro de Investigación Biomédica en Red en Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, Madrid, Spain.
- 13 Intensive Care Unit, Hospital Universitario La Princesa, Madrid, Spain.
- 14 Universidad Autónoma de Madrid (UAM), Madrid, Spain.
- PMID: 37809329
- PMCID: PMC10557488
- DOI: 10.3389/fmed.2023.1215246
Abstract
Introduction: SARS-CoV-2 viral load has been related to COVID-19 severity. The main aim of this study was to evaluate the relationship between SARS-CoV-2 viremia and SNPs in genes previously studied by our group as predictors of COVID-19 severity.
Materials and methods: Retrospective observational study including 340 patients hospitalized for COVID-19 in the University Hospital La Princesa between March 2020 and December 2021, with at least one viremia determination. Positive viremia was considered when viral load was above the quantifiable threshold (20 copies/ml). A total of 38 SNPs were genotyped. To study their association with viremia a multivariate logistic regression was performed.
Results: The mean age of the studied population was 64.5 years (SD 16.6), 60.9% patients were male and 79.4% white non-Hispanic. Only 126 patients (37.1%) had at least one positive viremia. After adjustment by confounders, the presence of the minor alleles of rs2071746 (HMOX1; T/T genotype OR 9.9 p < 0.0001), rs78958998 (probably associated with SERPING1 expression; A/T genotype OR 2.3, p = 0.04 and T/T genotype OR 12.9, p < 0.0001), and rs713400 (eQTL for TMPRSS2; C/T + T/T genotype OR 1.86, p = 0.10) were associated with higher risk of viremia, whereas the minor alleles of rs11052877 (CD69; A/G genotype OR 0.5, p = 0.04 and G/G genotype OR 0.3, p = 0.01), rs2660 (OAS1; A/G genotype OR 0.6, p = 0.08), rs896 (VIPR1; T/T genotype OR 0.4, p = 0.02) and rs33980500 (TRAF3IP2; C/T + T/T genotype OR 0.3, p = 0.01) were associated with lower risk of viremia.
Conclusion: Genetic variants in HMOX1 (rs2071746), SERPING1 (rs78958998), TMPRSS2 (rs713400), CD69 (rs11052877), TRAF3IP2 (rs33980500), OAS1 (rs2660) and VIPR1 (rs896) could explain heterogeneity in SARS-CoV-2 viremia in our population.
Keywords: COVID-19; SARS-CoV-2; genetic variants; single nucleotide polymorphism (SNPs); viremia.
Copyright © 2023 Roy-Vallejo, Fernández de Córdoba-Oñate, Delgado-Wicke, Triguero-Martínez, Montes, Carracedo-Rodríguez, Zurita-Cruz, Marcos-Jiménez, Lamana, Galván-Román, Villapalos García, Zubiaur, Ciudad, Rabes, Sanz, Rodríguez, Villa, Rodríguez, Marcos, Hernando, Díaz-Fernández, Abad, de los Santos, Rodríguez Serrano, García-Vicuña, Suárez Fernández, P. Gomariz, Muñoz-Calleja, Fernández-Ruiz, González-Álvaro Cardeñoso and the PREDINMUN-COVID Group.
Conflict of interest statement
FA, has been consultant or investigator in clinical trials sponsored by the following pharmaceutical companies: Abbott, Alter, Aptatargets, Chemo, FAES, Farmalíder, Ferrer, Galenicum, GlaxoSmithKline, Gilead, Italfarmaco, Janssen-Cilag, Kern, Normon, Novartis, Servier, Teva and Zambon. IG-Á reports grants from Instituto de Salud Carlos III, during the course of the study; personal fees from Lilly and Sanofi; personal fees and non-financial support from BMS; personal fees and non-financial support from Abbvie; research support, personal fees and non-financial support from Roche Laboratories; research support from Gebro Pharma; non-financial support from MSD, Pfizer and Novartis, not related to the submitted work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
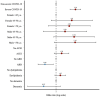
Figure 1
Clinical model. Forest plot with…
Figure 1
Clinical model. Forest plot with the Odds ratio and 95% Confidence Interval of…
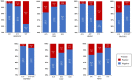
Figure 2
Predicted probability of viremia. Percentage…
Figure 2
Predicted probability of viremia. Percentage and 95% Confidence Interval of predicted probability of…
References
-
- World Health Organization . WHO coronavirus (COVID-19) dashboard. (2023) Available at: https://covid19.who.int/table. Accessed April 17, 2023
-
- Rodríguez-Serrano DA, Roy-Vallejo E, Zurita Cruz ND, Martín Ramírez A, Rodríguez-García SC, Arevalillo-Fernández N, et al. . Detection of SARS-CoV-2 RNA in serum is associated with increased mortality risk in hospitalized COVID-19 patients. Sci Rep. (2021) 11:13134. doi: 10.1038/s41598-021-92497-1, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

