A myeloid program associated with COVID-19 severity is decreased by therapeutic blockade of IL-6 signaling
- PMID: 37810211
- PMCID: PMC10551843
- DOI: 10.1016/j.isci.2023.107813
A myeloid program associated with COVID-19 severity is decreased by therapeutic blockade of IL-6 signaling
Abstract
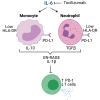
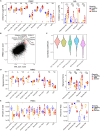
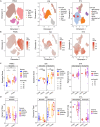
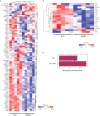
Altered myeloid inflammation and lymphopenia are hallmarks of severe infections. We identified the upregulated EN-RAGE gene program in airway and blood myeloid cells from patients with acute lung injury from SARS-CoV-2 or other causes across 7 cohorts. This program was associated with greater clinical severity and predicted future mechanical ventilation and death. EN-RAGEhi myeloid cells express features consistent with suppressor cell functionality, including low HLA-DR and high PD-L1. Sustained EN-RAGE program expression in airway and blood myeloid cells correlated with clinical severity and increasing expression of T cell dysfunction markers. IL-6 upregulated many EN-RAGE program genes in monocytes in vitro. IL-6 signaling blockade by tocilizumab in a placebo-controlled clinical trial led to rapid normalization of EN-RAGE and T cell gene expression. This identifies IL-6 as a key driver of myeloid dysregulation associated with worse clinical outcomes in COVID-19 patients and provides insights into shared pathophysiological mechanisms in non-COVID-19 ARDS.
Keywords: Clinical genetics; Immune response; Molecular medicine.
© 2023 The Authors.
Conflict of interest statement
J.A.H., H.S., J.V.H., C.O., L.O., X.G., N.W., A.Q., D.C., A.C, D.F.C., T.R., J.M.M., F.C., A.T., M.B., L.T., A.R., S.B.K., R.N.B., and C.M.R. were employees of Genentech, Inc. at the time of this study and own equity in Roche. The COMET study was supported in part by Genentech funding. C.J.Y. is a scientific advisory board member for and holds equity in Related Sciences and ImmunAI, a consultant for and holds equity in Maze Therapeutics, and a consultant for TRex Bio. C.J.Y. has received research support from Chan Zuckerberg Initiative, Chan Zuckerberg Biohub, and Genentech. C.S.C. has received research funding from Roche-Genentech for an unrelated project as well as from NIH, DOD, and Quantum Leap Healthcare Collaborative. C.S.C. is a consultant for Vasomune, Quark, and GEn1E Lifesciences. C.H. is a consultant for Spring Discovery but does not have any financial interest in the company nor is the work related to what is covered in this manuscript. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics and, until 31 July 2020, was a scientific advisory board member of Thermo Fisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. A.R. is a named inventor on multiple patents related to single-cell and spatial genomics filed by or issued to the Broad Institute.
Figures



References
-
- Zheng H., Rao A.M., Dermadi D., Toh J., Murphy Jones L., Donato M., Liu Y., Su Y., Dai C.L., Kornilov S.A., et al. Multi-cohort analysis of host immune response identifies conserved protective and detrimental modules associated with severity across viruses. Immunity. 2021;54:753–768.e5. doi: 10.1016/j.immuni.2021.03.002. - DOI - PMC - PubMed
-
- MacDonald L., Alivernini S., Tolusso B., Elmesmari A., Somma D., Perniola S., Paglionico A., Petricca L., Bosello S.L., Carfì A., et al. COVID-19 and RA share an SPP1 myeloid pathway that drives PD-L1+ neutrophils and CD14+ monocytes. JCI Insight. 2021;6 doi: 10.1172/jci.insight.147413. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

