Alternative splicing of BCL-x is controlled by RBM25 binding to a G-quadruplex in BCL-x pre-mRNA
- PMID: 37811881
- PMCID: PMC10639069
- DOI: 10.1093/nar/gkad772
Alternative splicing of BCL-x is controlled by RBM25 binding to a G-quadruplex in BCL-x pre-mRNA
Abstract
BCL-x is a master regulator of apoptosis whose pre-mRNA is alternatively spliced into either a long (canonical) anti-apoptotic Bcl-xL isoform, or a short (alternative) pro-apoptotic Bcl-xS isoform. The balance between these two antagonistic isoforms is tightly regulated and overexpression of Bcl-xL has been linked to resistance to chemotherapy in several cancers, whereas overexpression of Bcl-xS is associated to some forms of diabetes and cardiac disorders. The splicing factor RBM25 controls alternative splicing of BCL-x: its overexpression favours the production of Bcl-xS, whereas its downregulation has the opposite effect. Here we show that RBM25 directly and specifically binds to GQ-2, an RNA G-quadruplex (rG4) of BCL-x pre-mRNA that forms at the vicinity of the alternative 5' splice site leading to the alternative Bcl-xS isoform. This RBM25/rG4 interaction is crucial for the production of Bcl-xS and depends on the RE (arginine-glutamate-rich) motif of RBM25, thus defining a new type of rG4-interacting domain. PhenDC3, a benchmark G4 ligand, enhances the binding of RBM25 to the GQ-2 rG4 of BCL-x pre-mRNA, thereby promoting the alternative pro-apoptotic Bcl-xS isoform and triggering apoptosis. Furthermore, the screening of a combinatorial library of 90 putative G4 ligands led to the identification of two original compounds, PhenDH8 and PhenDH9, superior to PhenDC3 in promoting the Bcl-xS isoform and apoptosis. Thus, favouring the interaction between RBM25 and the GQ-2 rG4 of BCL-x pre-mRNA represents a relevant intervention point to re-sensitize cancer cells to chemotherapy.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


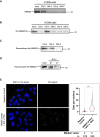

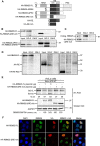

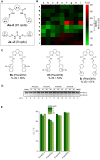
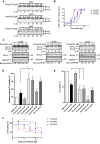
References
-
- Chow L.T., Gelinas R.E., Broker T.R., Roberts R.J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977; 12:1–8. - PubMed
-
- Chow L.T., Roberts J.M., Lewis J.B., Broker T.R. A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids. Cell. 1977; 11:819–836. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

