OvaPrint-A Cell-free DNA Methylation Liquid Biopsy for the Risk Assessment of High-grade Serous Ovarian Cancer
- PMID: 37812492
- PMCID: PMC10722131
- DOI: 10.1158/1078-0432.CCR-23-1197
OvaPrint-A Cell-free DNA Methylation Liquid Biopsy for the Risk Assessment of High-grade Serous Ovarian Cancer
Abstract
Purpose: High-grade serous ovarian carcinoma (HGSOC) is the most lethal epithelial ovarian cancer (EOC) and is often diagnosed at late stage. In women with a known pelvic mass, surgery followed by pathologic assessment is the most reliable way to diagnose EOC and there are still no effective screening tools in asymptomatic women. In the current study, we developed a cell-free DNA (cfDNA) methylation liquid biopsy for the risk assessment of early-stage HGSOC.
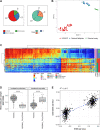
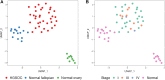
Experimental design: We performed reduced representation bisulfite sequencing to identify differentially methylated regions (DMR) between HGSOC and normal ovarian and fallopian tube tissue. Next, we performed hybridization probe capture for 1,677 DMRs and constructed a classifier (OvaPrint) on an independent set of cfDNA samples to discriminate HGSOC from benign masses. We also analyzed a series of non-HGSOC EOC, including low-grade and borderline samples to assess the generalizability of OvaPrint. A total of 372 samples (tissue n = 59, plasma n = 313) were analyzed in this study.
Results: OvaPrint achieved a positive predictive value of 95% and a negative predictive value of 88% for discriminating HGSOC from benign masses, surpassing other commercial tests. OvaPrint was less sensitive for non-HGSOC EOC, albeit it may have potential utility for identifying low-grade and borderline tumors with higher malignant potential.
Conclusions: OvaPrint is a highly sensitive and specific test that can be used for the risk assessment of HGSOC in symptomatic women. Prospective studies are warranted to validate OvaPrint for HGSOC and further develop it for non-HGSOC EOC histotypes in both symptomatic and asymptomatic women with adnexal masses.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet 2019;393:1240–53. - PubMed
-
- Surveillance, Epidemiology, and End Results (SEER) Program.SEER*Stat Database: Incidence - SEER 9 Regs Research Data, Nov 2018 Sub (1975–2016) <Katrina/Rita Population Adjustment>. Available from: https://www.seer.cancer.gov/.
-
- MacLaughlan S, Cronin B, Moore RG. Evaluation and management of women presenting with a pelvic mass. Curr Obstet Gynecol Rep 2012;1:10–5.
-
- Biggs WS, Marks ST. Diagnosis and management of adnexal masses. Am Fam Physician 2016;93:676–81. - PubMed
-
- Givens V, Mitchell GE, Harraway-Smith C, Reddy A, Maness DL. Diagnosis and management of adnexal masses. Am Fam Physician 2009;80:815–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

