Phenotype and fate of liver-resident CD8 T cells during acute and chronic hepacivirus infection
- PMID: 37812637
- PMCID: PMC10602381
- DOI: 10.1371/journal.ppat.1011697
Phenotype and fate of liver-resident CD8 T cells during acute and chronic hepacivirus infection
Abstract
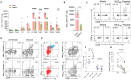
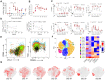
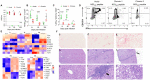
Immune correlates of hepatitis C virus (HCV) clearance and control remain poorly defined due to the lack of an informative animal model. We recently described acute and chronic rodent HCV-like virus (RHV) infections in lab mice. Here, we developed MHC class I and class II tetramers to characterize the serial changes in RHV-specific CD8 and CD4 T cells during acute and chronic infection in C57BL/6J mice. RHV infection induced rapid expansion of T cells targeting viral structural and nonstructural proteins. After virus clearance, the virus-specific T cells transitioned from effectors to long-lived liver-resident memory T cells (TRM). The effector and memory CD8 and CD4 T cells primarily produced Th1 cytokines, IFN-γ, TNF-α, and IL-2, upon ex vivo antigen stimulation, and their phenotype and transcriptome differed significantly between the liver and spleen. Rapid clearance of RHV reinfection coincided with the proliferation of virus-specific CD8 TRM cells in the liver. Chronic RHV infection was associated with the exhaustion of CD8 T cells (Tex) and the development of severe liver diseases. Interestingly, the virus-specific CD8 Tex cells continued proliferation in the liver despite the persistent high-titer viremia and retained partial antiviral functions, as evident from their ability to degranulate and produce IFN-γ upon ex vivo antigen stimulation. Thus, RHV infection in mice provides a unique model to study the function and fate of liver-resident T cells during acute and chronic hepatotropic infection.
Copyright: © 2023 Dravid et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Zibbell JE, Asher AK, Patel RC, Kupronis B, Iqbal K, Ward JW, et al.. Increases in Acute Hepatitis C Virus Infection Related to a Growing Opioid Epidemic and Associated Injection Drug Use, United States, 2004 to 2014. American journal of public health. 2018;108(2):175–81. doi: 10.2105/AJPH.2017.304132 - DOI - PMC - PubMed
-
- Bartenschlager R, Baumert TF, Bukh J, Houghton M, Lemon SM, Lindenbach BD, et al.. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: Considerations for scientists and funding agencies. Virus Res. 2018;248:53–62. doi: 10.1016/j.virusres.2018.02.016 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

