Single Nucleotide Polymorphisms Associated With Motor Recovery in Patients With Nondisabling Stroke: GWAS Study
- PMID: 37813584
- PMCID: PMC10663021
- DOI: 10.1212/WNL.0000000000207716
Single Nucleotide Polymorphisms Associated With Motor Recovery in Patients With Nondisabling Stroke: GWAS Study
Erratum in
-
Corrections to Preprint Server Information.Neurology. 2024 Jul 9;103(1):e209573. doi: 10.1212/WNL.0000000000209573. Epub 2024 Jun 3. Neurology. 2024. PMID: 38830142 Free PMC article. No abstract available.
Abstract
Background and objectives: Despite notable advances in genetic understanding of stroke recovery, most studies focus only on candidate genes. To date, only 2 genome-wide association studies (GWAS) have focused on stroke outcomes, but they were limited to the modified Rankin Scale (mRS). The mRS maps poorly to biological processes. Therefore, we performed a GWAS to discover single nucleotide polymorphisms (SNPs) associated with motor recovery poststroke.
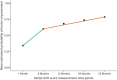
Methods: We used the Vitamin Intervention for Stroke Prevention (VISP) data set of 2,100 genotyped participants with nondisabling stroke. We included only participants who had motor impairment at randomization. Participants with a recurrent stroke during the trial were excluded. Genotyped data underwent strict quality control and imputation. The GWAS used logistic regression models with generalized estimating equations to leverage the repeated NIH Stroke Scale motor score measurements spanning 6 time points over 24 months. The primary outcome was a decrease in the motor drift score of ≥1 vs <1 at each time point. Our model estimated the odds ratio (OR) of motor improvement for each SNP after adjusting for age, sex, race, days from stroke to visit, initial motor score, VISP treatment arm, and principal components.
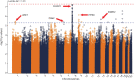
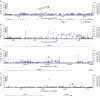
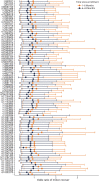
Results: A total of 488 (64%) participants with a mean (SD) age of 66 ± 11 years were included in the GWAS. Although no associations reached genome-wide significance (p < 5 × 10-8), our analysis detected 115 suggestive associations (p < 5 × 10-6). Notably, we found multiple SNP clusters near genes with plausible neuronal repair biology mechanisms. The CLDN23 gene had the most convincing association with rs1268196-T as its most significant SNP (OR 0.32; 95% CI 0.21-0.48; p value 6.19 × 10-7). CLDN23 affects blood-brain barrier integrity, neurodevelopment, and immune cell transmigration.
Discussion: We identified novel suggestive genetic associations with the first-ever motor-specific poststroke recovery GWAS. The results seem to describe a distinct stroke recovery phenotype compared with prior genetic stroke outcome studies that use outcome measures, such as the mRS. Replication and further mechanistic investigation are warranted. In addition, this study demonstrated a proof-of-principle approach to optimize statistical efficiency with longitudinal data sets for genetic discovery.
© 2023 American Academy of Neurology.
Conflict of interest statement
B.B. Worrall serves as the Deputy Editor for the journal Neurology. All other authors report no relevant disclosures. Go to
Figures

References
Publication types
MeSH terms
Grants and funding
- U01 HG005160/HG/NHGRI NIH HHS/United States
- U01 HG005157/HG/NHGRI NIH HHS/United States
- R01 NS034447/NS/NINDS NIH HHS/United States
- R01 NS034447/NS/NINDS NIH HHS/United States
- R01 CA100264/CA/NCI NIH HHS/United States
- P50 CA093459/CA/NCI NIH HHS/United States
- P50 CA097007/CA/NCI NIH HHS/United States
- R01 ES011740/ES/NIEHS NIH HHS/United States
- R01 ES011740/ES/NIEHS NIH HHS/United States
- R01 CA133996/CA/NCI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- Z01 AG000513/ImNIH/Intramural NIH HHS/United States
- ZIA AG000513/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
