GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
- PMID: 37813859
- PMCID: PMC10562414
- DOI: 10.1038/s41467-023-41893-4
GLP-1R signaling neighborhoods associate with the susceptibility to adverse drug reactions of incretin mimetics
Abstract
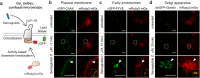
G protein-coupled receptors are important drug targets that engage and activate signaling transducers in multiple cellular compartments. Delineating therapeutic signaling from signaling associated with adverse events is an important step towards rational drug design. The glucagon-like peptide-1 receptor (GLP-1R) is a validated target for the treatment of diabetes and obesity, but drugs that target this receptor are a frequent cause of adverse events. Using recently developed biosensors, we explored the ability of GLP-1R to activate 15 pathways in 4 cellular compartments and demonstrate that modifications aimed at improving the therapeutic potential of GLP-1R agonists greatly influence compound efficacy, potency, and safety in a pathway- and compartment-selective manner. These findings, together with comparative structure analysis, time-lapse microscopy, and phosphoproteomics, reveal unique signaling signatures for GLP-1R agonists at the level of receptor conformation, functional selectivity, and location bias, thus associating signaling neighborhoods with functionally distinct cellular outcomes and clinical consequences.
© 2023. Springer Nature Limited.
Conflict of interest statement
M.B. is the president of the scientific advisory board for Domain Therapeutics. M.B. and C.L.G. have filed patent application (US20200256869A1) related to some of the biosensors used in this work and the technology has been licensed to Domain Therapeutics. All biosensors are available for academic research through regular material transfer agreement. V.M.L. is co-founder, the CEO, and shareholder of HepaPredict AB, as well as co-founder and shareholder of PersoMedix AB. A.M., S.K., P.S., C.M.B., A.B., E.B.-T., K.M., I.P., D-C.S., N.A.L., J.V.O., R.A.Z., A.S.H. and S.C.W. declare no competing interests.
Figures


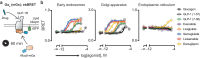


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

