Fibroblast growth factor 18 stimulates the proliferation of hepatic stellate cells, thereby inducing liver fibrosis
- PMID: 37813881
- PMCID: PMC10562492
- DOI: 10.1038/s41467-023-42058-z
Fibroblast growth factor 18 stimulates the proliferation of hepatic stellate cells, thereby inducing liver fibrosis
Abstract
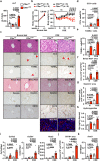
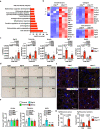
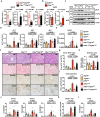
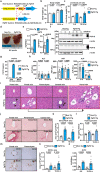
Liver fibrosis results from chronic liver injury triggered by factors such as viral infection, excess alcohol intake, and lipid accumulation. However, the mechanisms underlying liver fibrosis are not fully understood. Here, we demonstrate that the expression of fibroblast growth factor 18 (Fgf18) is elevated in mouse livers following the induction of chronic liver fibrosis models. Deletion of Fgf18 in hepatocytes attenuates liver fibrosis; conversely, overexpression of Fgf18 promotes liver fibrosis. Single-cell RNA sequencing reveals that overexpression of Fgf18 in hepatocytes results in an increase in the number of Lrat+ hepatic stellate cells (HSCs), thereby inducing fibrosis. Mechanistically, FGF18 stimulates the proliferation of HSCs by inducing the expression of Ccnd1. Moreover, the expression of FGF18 is correlated with the expression of profibrotic genes, such as COL1A1 and ACTA2, in human liver biopsy samples. Thus, FGF18 promotes liver fibrosis and could serve as a therapeutic target to treat liver fibrosis.
© 2023. Springer Nature Limited.
Conflict of interest statement
Y.T. and Hiroyasu N. are inventors on the patent application that includes the ELISA to detect human and murine FGF18 and the generation of transgenic mice that spontaneously developed liver fibrosis used in this study. The other authors declare no conflicts of interest.
Figures



References
-
- Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 2021;18:151–166. - PubMed
-
- Vilar-Gomez E, et al. Fibrosis severity as a determinant of cause-specific mortality in patients with advanced nonalcoholic fatty liver disease: a multi-national cohort study. Gastroenterology. 2018;155:443–457.e417. - PubMed
-
- Rinella ME, et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023;S0168–8278:00418–X. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

