ctDNA response after pembrolizumab in non-small cell lung cancer: phase 2 adaptive trial results
- PMID: 37814061
- PMCID: PMC10579094
- DOI: 10.1038/s41591-023-02598-9
ctDNA response after pembrolizumab in non-small cell lung cancer: phase 2 adaptive trial results
Abstract
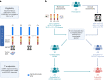
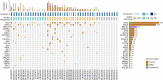
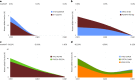
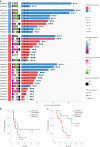
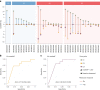
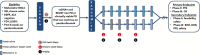
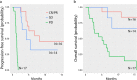
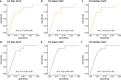
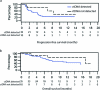
Circulating tumor DNA (ctDNA) has shown promise in capturing primary resistance to immunotherapy. BR.36 is a multi-center, randomized, ctDNA-directed, phase 2 trial of molecular response-adaptive immuno-chemotherapy for patients with lung cancer. In the first of two independent stages, 50 patients with advanced non-small cell lung cancer received pembrolizumab as standard of care. The primary objectives of stage 1 were to ascertain ctDNA response and determine optimal timing and concordance with radiologic Response Evaluation Criteria in Solid Tumors (RECIST) response. Secondary endpoints included the evaluation of time to ctDNA response and correlation with progression-free and overall survival. Maximal mutant allele fraction clearance at the third cycle of pembrolizumab signified molecular response (mR). The trial met its primary endpoint, with a sensitivity of ctDNA response for RECIST response of 82% (90% confidence interval (CI): 52-97%) and a specificity of 75% (90% CI: 56.5-88.5%). Median time to ctDNA response was 2.1 months (90% CI: 1.5-2.6), and patients with mR attained longer progression-free survival (5.03 months versus 2.6 months) and overall survival (not reached versus 7.23 months). These findings are incorporated into the ctDNA-driven interventional molecular response-adaptive second stage of the BR.36 trial in which patients at risk of progression are randomized to treatment intensification or continuation of therapy. ClinicalTrials.gov ID: NCT04093167 .
© 2023. The Author(s).
Conflict of interest statement
V.A. receives research funding to Johns Hopkins University from AstraZeneca and Personal Genome Diagnostics; has received research funding to Johns Hopkins University from Bristol-Myers Squibb and Delfi Diagnostics in the past 5 years; and is an advisory board member for AstraZeneca and NeoGenomics. V.A is an inventor on patent applications (63/276,525, 17/779,936, 16/312,152, 16/341,862, 17/047,006 and 17/598,690) submitted by Johns Hopkins University related to cancer genomic analyses, ctDNA therapeutic response monitoring and immunogenomic features of response to immunotherapy, which have been licensed to one or more entities. Under the terms of these license agreements, the university and inventors are entitled to fees and royalty distributions. N.N. is an inventor on patent application 17/598,690 submitted by Johns Hopkins University related to genomic features of response to immunotherapy. S.L. receives research funding to his institution from Bristol-Myers Squibb, Merck, GlaxoSmithKline, 23andme, SignalChem, AstraZeneca, Treadwell Therapeutics and Boehringer Ingelheim; serves on the advisory boards of Sanofi, Bayer and Pfizer; and holds royalties from UpToDate. M.S. is an employee of Personal Genome Diagnostics (LabCorp) and holds equity in LabCorp. R.A.J. has participated on advisory boards for AbbVie, Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, EMD Serono, Fusion Pharmaceuticals, Jazz Pharmaceuticals, Eli Lilly, Merck Sharp & Dohme, Novartis, Pfizer, Roche Canada, Sanofi/Regeneron and Takeda; has received honoraria from Amgen, AstraZeneca, Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis Pharmaceuticals Canada and Roche Canada; and has received research funding from AstraZeneca/MedImmune, Bristol-Myers Squibb, Debiopharm Group, Merck Sharp & Dohme, Novartis and Turnstone Bio. A.S.F. receives research funding from AstraZeneca and has served on advisory boards for Novartis. J.B. receives institutional grant support from Bristol-Myers Squibb and AstraZeneca; is an advisory board member for Bristol-Myers Squibb, Merck, AstraZeneca and Regeneron; and serves as a Data and Safety Monitoring Board member for Sanofi, GlaxoSmithKline and Johnson & Johnson. P.O.G. reports research/clinical trial funding from Stand Up to Cancer Canada–Canadian Cancer Society Breast Cancer Dream Team, AstraZeneca, Merck, BioAtla, Novartis and the Cancer Research Institute. A.S. reports research funding from Genentech/Roche, AstraZeneca and Bristol-Myers Squibb and serves as a consultant for Amgen, AstraZeneca and Genentech/Roche. P.A.B. has participated on advisory boards for Abbvie, Mirati, AstraZeneca and Boehringer Ingelheim and has received honoraria from Merck, Pfizer and Eli Lilly. B.L. is a consultant for AstraZeneca, Daiichi Sankyo, Janssen, Bristol-Myers Squibb, Novartis, Genentech, Eli Lilly, Pfizer, Guardant 360, Mirati, Amgen and Foundation One. J.D. reports research grants to her institution from Pfizer, Merck, Bristol-Myers Squibb, AstraZeneca, Seagen and Inivata. All other authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

