Claudin-2 protects against colitis-associated cancer by promoting colitis-associated mucosal healing
- PMID: 37815870
- PMCID: PMC10688979
- DOI: 10.1172/JCI170771
Claudin-2 protects against colitis-associated cancer by promoting colitis-associated mucosal healing
Abstract
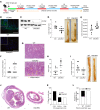
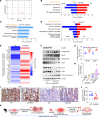
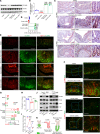
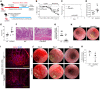
Patients with inflammatory bowel disease (IBD) are susceptible to colitis-associated cancer (CAC). Chronic inflammation promotes the risk for CAC. In contrast, mucosal healing predicts improved prognosis in IBD and reduced risk of CAC. However, the molecular integration among colitis, mucosal healing, and CAC remains poorly understood. Claudin-2 (CLDN2) expression is upregulated in IBD; however, its role in CAC is not known. The current study was undertaken to examine the role for CLDN2 in CAC. The AOM/DSS-induced CAC model was used with WT and CLDN2-modified mice. High-throughput expression analyses, murine models of colitis/recovery, chronic colitis, ex vivo crypt culture, and pharmacological manipulations were employed in order to increase our mechanistic understanding. The Cldn2KO mice showed significant inhibition of CAC despite severe colitis compared with WT littermates. Cldn2 loss also resulted in impaired recovery from colitis and increased injury when mice were subjected to intestinal injury by other methods. Mechanistic studies demonstrated a possibly novel role of CLDN2 in promotion of mucosal healing downstream of EGFR signaling and by regulation of Survivin expression. An upregulated CLDN2 expression protected from CAC and associated positively with crypt regeneration and Survivin expression in patients with IBD. We demonstrate a potentially novel role of CLDN2 in promotion of mucosal healing in patients with IBD and thus regulation of vulnerability to colitis severity and CAC, which can be exploited for improved clinical management.
Keywords: Colorectal cancer; Gastroenterology; Inflammatory bowel disease; Tight junctions.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

