Hepatitis B virus infection disrupts homologous recombination in hepatocellular carcinoma by stabilizing resection inhibitor ADRM1
- PMID: 37815873
- PMCID: PMC10688980
- DOI: 10.1172/JCI171533
Hepatitis B virus infection disrupts homologous recombination in hepatocellular carcinoma by stabilizing resection inhibitor ADRM1
Abstract
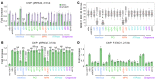
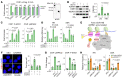
Many cancers harbor homologous recombination defects (HRDs). A HRD is a therapeutic target that is being successfully utilized in treatment of breast/ovarian cancer via synthetic lethality. However, canonical HRD caused by BRCAness mutations do not prevail in liver cancer. Here we report a subtype of HRD caused by the perturbation of a proteasome variant (CDW19S) in hepatitis B virus-bearing (HBV-bearing) cells. This amalgamate protein complex contained the 19S proteasome decorated with CRL4WDR70 ubiquitin ligase, and assembled at broken chromatin in a PSMD4Rpn10- and ATM-MDC1-RNF8-dependent manner. CDW19S promoted DNA end processing via segregated modules that promote nuclease activities of MRE11 and EXO1. Contrarily, a proteasomal component, ADRM1Rpn13, inhibited resection and was removed by CRL4WDR70-catalyzed ubiquitination upon commitment of extensive resection. HBx interfered with ADRM1Rpn13 degradation, leading to the imposition of ADRM1Rpn13-dependent resection barrier and consequent viral HRD subtype distinguishable from that caused by BRCA1 defect. Finally, we demonstrated that viral HRD in HBV-associated hepatocellular carcinoma can be exploited to restrict tumor progression. Our work clarifies the underlying mechanism of a virus-induced HRD subtype.
Keywords: DNA repair; Hepatology; Liver cancer; Ubiquitin-proteosome system; Virology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

