Insulin-degrading enzyme (IDE) as a modulator of microglial phenotypes in the context of Alzheimer's disease and brain aging
- PMID: 37817156
- PMCID: PMC10566021
- DOI: 10.1186/s12974-023-02914-7
Insulin-degrading enzyme (IDE) as a modulator of microglial phenotypes in the context of Alzheimer's disease and brain aging
Abstract
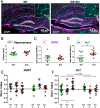
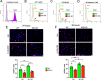
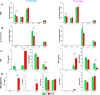
The insulin-degrading enzyme (IDE) is an evolutionarily conserved zinc-dependent metallopeptidase highly expressed in the brain, where its specific functions remain poorly understood. Besides insulin, IDE is able to cleave many substrates in vitro, including amyloid beta peptides, making this enzyme a candidate pathophysiological link between Alzheimer's disease (AD) and type 2 diabetes (T2D). These antecedents led us to address the impact of IDE absence in hippocampus and olfactory bulb. A specific induction of microgliosis was found in the hippocampus of IDE knockout (IDE-KO) mice, without any effects in neither hippocampal volume nor astrogliosis. Performance on hippocampal-dependent memory tests is influenced by IDE gene dose in 12-month-old mice. Furthermore, a comprehensive characterization of the impact of IDE haploinsufficiency and total deletion in metabolic, behavioral, and molecular parameters in the olfactory bulb, a site of high insulin receptor levels, reveals an unambiguous barcode for IDE-KO mice at that age. Using wildtype and IDE-KO primary microglial cultures, we performed a functional analysis at the cellular level. IDE absence alters microglial responses to environmental signals, resulting in impaired modulation of phenotypic states, with only transitory effects on amyloid-β management. Collectively, our results reveal previously unknown physiological functions for IDE in microglia that, due to cell-compartment topological reasons, cannot be explained by its enzymatic activity, but instead modulate their multidimensional response to various damaging conditions relevant to aging and AD conditions.
Keywords: Amyloid-beta endocytosis; Cytokine secretion; Inflammation; Insulin-degrading enzyme; Microglia; Microglial proliferation; Myelin phagocytosis; Oxidative stress.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures



References
-
- Nakabeppu Y, Ninomiya T. Diabetes mellitus: a risk factor for Alzheimer’s disease. Singapore: Springer; 2019.
MeSH terms
Substances
Grants and funding
- "Margarita Salas postdoctoral grant for the training of young doctors"/Ministerio de Universidades
- Predoctoral fellowship/Universidad de Valladolid
- PID2019-110496RB-C21/Ministerio de Ciencia e Innovación
- PID2019-110911RB-I00/AEI/Ministerio de Ciencia e Innovación
- VA086G18/Consejería de Educación, Junta de Castilla y León
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

