The GmSTF1/2-GmBBX4 negative feedback loop acts downstream of blue-light photoreceptors to regulate isoflavonoid biosynthesis in soybean
- PMID: 37817409
- PMCID: PMC10873893
- DOI: 10.1016/j.xplc.2023.100730
The GmSTF1/2-GmBBX4 negative feedback loop acts downstream of blue-light photoreceptors to regulate isoflavonoid biosynthesis in soybean
Abstract
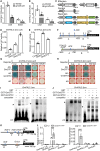
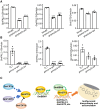
Isoflavonoids, secondary metabolites derived from the phenylalanine pathway, are predominantly biosynthesized in legumes, especially soybean (Glycine max). They are not only essential for plant responses to biotic and abiotic stresses but also beneficial to human health. In this study, we report that light signaling controls isoflavonoid biosynthesis in soybean. Blue-light photoreceptors (GmCRY1s, GmCRY2s, GmPHOT1s, and GmPHOT2s) and the transcription factors GmSTF1 and GmSTF2 promote isoflavonoid accumulation, whereas the E3 ubiquitin ligase GmCOP1b negatively regulates isoflavonoid biosynthesis. GmPHOT1s and GmPHOT2s stabilize GmSTF1/2, whereas GmCOP1b promotes the degradation of these two proteins in soybean. GmSTF1/2 regulate the expression of approximately 27.9% of the genes involved in soybean isoflavonoid biosynthesis, including GmPAL2.1, GmPAL2.3, and GmUGT2. They also repress the expression of GmBBX4, a negative regulator of isoflavonoid biosynthesis in soybean. In addition, GmBBX4 physically interacts with GmSTF1 and GmSTF2 to inhibit their transcriptional activation activity toward target genes related to isoflavonoid biosynthesis. Thus, GmSTF1/2 and GmBBX4 form a negative feedback loop that acts downstream of photoreceptors in the regulation of isoflavonoid biosynthesis. Our study provides novel insights into the control of isoflavonoid biosynthesis by light signaling in soybean and will contribute to the breeding of soybean cultivars with high isoflavonoid content through genetic and metabolic engineering.
Keywords: GmBBX4; GmSTF; isoflavonoid; light signaling; photoreceptor; soybean.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Alba R., Kelmenson P.M., Cordonnier-Pratt M.M., Pratt L.H. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 2000;17:362–373. - PubMed
-
- Araguirang G.E., Richter A.S. Activation of anthocyanin biosynthesis in high light - what is the initial signal? New Phytol. 2022;236:2037–2043. - PubMed
-
- Bai S., Tao R., Yin L., Ni J., Yang Q., Yan X., Yang F., Guo X., Li H., Teng Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019;100:1208–1223. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

