Tetracyclines activate mitoribosome quality control and reduce ER stress to promote cell survival
- PMID: 37818824
- PMCID: PMC10702820
- DOI: 10.15252/embr.202357228
Tetracyclines activate mitoribosome quality control and reduce ER stress to promote cell survival
Abstract
Mitochondrial diseases are a group of disorders defined by defects in oxidative phosphorylation caused by nuclear- or mitochondrial-encoded gene mutations. A main cellular phenotype of mitochondrial disease mutations is redox imbalances and inflammatory signaling underlying pathogenic signatures of these patients. One method to rescue this cell death vulnerability is the inhibition of mitochondrial translation using tetracyclines. However, the mechanisms whereby tetracyclines promote cell survival are unknown. Here, we show that tetracyclines inhibit the mitochondrial ribosome and promote survival through suppression of endoplasmic reticulum (ER) stress. Tetracyclines increase mitochondrial levels of the mitoribosome quality control factor MALSU1 (Mitochondrial Assembly of Ribosomal Large Subunit 1) and promote its recruitment to the mitoribosome large subunit, where MALSU1 is necessary for tetracycline-induced survival and suppression of ER stress. Glucose starvation induces ER stress to activate the unfolded protein response and IRE1α-mediated cell death that is inhibited by tetracyclines. These studies establish a new interorganelle communication whereby inhibition of the mitoribosome signals to the ER to promote survival, implicating basic mechanisms of cell survival and treatment of mitochondrial diseases.
Keywords: IRE1α; MALSU1; mitochondrial disease; mitoribosome; tetracyclines.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

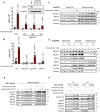
ND1 cells treated with a range of concentrations (0.1, 1.0, 10, 100 μM) of peptide deformylase inhibitor actinonin do not promote survival under galactose conditions (error bars represent the average ± s.e.m. of n = 3 independent biological samples). Statistical significance (P < 0.05) between actinonin and untreated cultures was calculated using one‐way ANOVA, multiple comparisons.
DepMap drug‐repurposing proteomics database indicates enrichment of mitochondrial and more specifically mitoribosome proteins in the context of doxycycline. Gene‐ontology enrichment was determined using DAVID pathway analysis, where significant proteins were determined (P < 0.001) and compared to entire proteome. Note enrichment of MALSU1 associated with surviving cells in the context of doxycycline. Statistical significance was determined as previously described (Huang da et al, 2009).
Depletion (CRISPR‐Cas9) of mitochondrial quality control rescue factors MTRES1 and mtRF‐R did not reverse capacity of tetracyclines to rescue ND1 cells from galactose induced cell death (error bars represent the average ± s.e.m. of n = 11 biological replicates over N = 4 experiments). During survival experiments, dashed‐line represents ND1 seeding density, where cells were exposed to glucose deprivation (galactose supplementation) for 4 days. P‐values* with asterisk denote comparisons with DMSO treated NTC cells. Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC, sgMTRES1, and sgmtRF‐R cells was calculated using two‐way ANOVA, multiple comparisons.
MALSU1 expression is required for tetracyclines induced survival (error bars represent the average ± s.e.m. of n = 6 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC and sgMALSU1 (sg1‐sg3 independent small‐guide RNA) was calculated using two‐way ANOVA, multiple comparisons.
The broad class of tetracycline antibiotics rescue ND1 cells from nutrient deprivation depending on MALSU1. Doxycycline, tetracycline, minocycline, and 7002 were utilized as structurally diverse tetracycline analogs that cannot rescue ND1 mutant cells that lack MALSU1 (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between tetracycline analogs and untreated cells across NTC and sgMALSU1 was calculated using two‐way ANOVA, multiple comparisons. Chemical modifications of each respective analog from parental tetracycline template are highlighted in red.
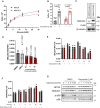
- A
CRISPR‐Cas9 KO of MTIF3 in ND1 cybrid cells reduces doxycycline's ability to promote survival (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC and sgMTIF3 (sg1‐sg3) was calculated using two‐way ANOVA, multiple comparisons.
- B
CRISPR‐Cas9 KO of MALSU1 and MTIF3 drastically reduce the ability of tetracyclines to promote survival when compared to LOR8F8‐KO (error bars represent the average ± s.e.m. of n = 6 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across NTC, sgMALSU1, sgLOR8F8, and sgMTIF3 cells was calculated using two‐way ANOVA, multiple comparisons.
- C
MALSU1 is recruited to the mitochondria at early time points. Crude mitochondria (mitochondria/ER) were isolated from Expi293F cells at varying time points following doxycycline treatment. Note maximum mitochondrial MALSU1 at the 12‐h time point.
- D
Mitoribosome isolation in Expi293F cells indicates doxycycline promotes the accumulation of MALSU1 staring at 1 h, with drastic increases at the 12‐h time point. Note mitochondrial accumulation of MALSU1 at 12 h. MTIF3 expression, along with mitochondrial and mitoribosome accumulation were unchanged with doxycycline.
- E, F
(E) Mitochondria were isolated from ND1 mutant cybrid cells exposed to galactose with or without doxycycline for varying time points, with rapid mitochondrial accumulation of MALSU1 after 1 h, that maximizes from 3 to 24 h of doxycycline treatment. Note subtle increases in MTIF3, along with MRPL4 and MRPL6 at the 3‐h time points. (F) Matched glucose controls at the 3‐ and 24‐h time point illustrate a decrease in mitochondrial MALSU1, MRPL4, and MRPS16 upon galactose treatment that is stabilized with doxycycline. Note increased whole cell expression level of MALSU1 in doxycycline treated cultures at the 24‐h time point.
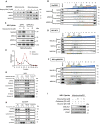
- A
ND1 control (MOCK) and MALSU1‐OE cells exposed to varying extent of glucose deprivation (96 h) follow similar cell death responses (error bars represent the average ± s.e.m. of n = 3 biological replicates per concentration).
- B
MALSU1‐OE ND1 cells do not synergize with doxycycline (1 μM) in promoting survival (96 h, error bars represent the average ± s.e.m. of n = 9 biological replicates across N = 3 independent experiments). Statistical significance (P < 0.05) between doxycycline treated and untreated cells across MOCK and MALSU1‐OE cells was calculated using two‐way ANOVA, multiple comparisons.
- C
Stable MALSU1‐flag tag expression in ND1 cells.
- D
Quantitative PCR of ND1 cells (galactose, 48 h) treated with varying concentrations of doxycycline or inactive tetracycline analog CMT3 indicate mRNA expression levels of MALSU1 are unchanged (error bars represent the average ± s.e.m. of n = 4 biological replicates). Statistical significance (P < 0.05) between doxycycline or CMT3 treated and untreated cells was calculated using one‐way ANOVA, multiple comparisons.
- E, F
Time‐course experiments indicate that doxycycline does not alter (E) MALSU1 or (F) MTIF3 transcript levels (n = 2 technical replicates).
- G
ND1 cells exposed to glucose deprivation for indicated time periods with and without doxycycline indicate stability of MALSU1 by doxycycline at the 24 h time point. Note MTIF3 expression levels are unchanged. Note doxycycline suppresses the phosphorylation of p38 MAPK at later time points (12–24 h) as a control.
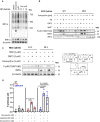
- A, B
(A) Isolated mitochondria and mitoribosomes from Expi293F exhibit enrichment of MALSU1 in doxycycline treated cultures, that is (B) associated with the large subunit of the mitoribosomes evidenced by sucrose gradient sedimentation (15–30% sucrose). Sedimentation of large and small subunit were assessed by MRPL4 and MRPS16, respectively. Duplicates of mitochondria and mitoribosomes in (A, n = 2 independent experiments) are the inputs for (B). Note shift in MALSU1 and MRPL4 upon doxycycline treatment.
- C
Isolation of crude mitochondria from doxycycline (1 μM) treated ND1 cells (non‐target control (NTC) and sgMALSU1). Note increased MALSU1 expression in doxycycline‐treated mitochondria.
- D
Crude mitochondria from ND1‐NTC cells were sucrose gradient (15–30%) sedimented and analyzed for MALSU1 as in (B). Note shift and enrichment of MALSU1 and MRPL4 with doxycycline treatment.
- E
Quantitation of MALSU1 elution profile in sucrose gradients of crude mitochondria from ND1 cells derived from (D), normalized to input signal of MALSU1.
- F
Sucrose gradient (15–30%) sedimentation of MALSU1‐depleted ND1 cells with or without doxycycline treatment. Crude mitochondria from (C) are inputs for (D) and (F).
- G, H
Cell fractionation of (G) ND1 and (H) Expi293F suspension cultures treated with doxycycline. Note enrichment of MALSU1 specifically in mitochondrial fractions and decreases in mitochondrial markers TOM70 and Cytochrome C at the whole cell level.
- I
ND1 cells treated with increasing concentrations of doxycycline illustrate dose‐dependent increases in MALSU1 specifically at the mitochondria without changes in total mitochondria (VDAC, LRPPRC).

- A
Doxycycline (1 μM) reverses activation of IRE1α as evidenced by decrease in XBP1s in ND1 cells (galactose, 48 and 72 h). Inactive CMT3 tetracycline analog does not reverse XBP1s. Suppression of XBP1s at 72 h is associated with reversed PARP cleavage.
- B
Blue‐Native PAGE indicates dose‐dependent reductions in high‐order IRE1α oligomers in isolated crude mitochondria (Mitochondria‐ER). Doxycycline dose‐dependently inhibits activation of XBP1s, ER chaperone BiP, and phosphorylation of p38 MAPK in ND1 cells (galactose, 48 h).
- C
IRE1α oligomerization status in mitochondria‐ER fractions and effector UPR function in ND1 cells is MALSU1 dependent. Note increased oligomerization of IRE1α upon galactose treatment that it attenuated in doxycycline treated cultures (non‐target control, NTC), with reductions in BiP and XBP1s. IRE1α oligomerization status was not attenuated by doxycycline in ND1 cells lacking MALSU1, and consistent with BiP and XBP1s.
- D
IRE1α expression levels in the brains of NDUFS4−/− mice indicate increased expression when compared to WT and doxycycline‐supplemented counterparts. Results represent whole brains isolated from three independent mice under each indicated condition (n = 3 independent biological replicates).
- E, F
Tetracyclines attenuate all three arms of the UPR. (E) Control (WT) and ND1 cybrids with (NTC) or without (sgMALSU1) MALSU1 were cultured in glucose, galactose (with or without doxycycline) for 48 h and analyzed for activation of IRE1α (phosphorylation), ATF6 (cleavage), and PERK (gel‐shift phosphorylation), along with expression levels of XBP1s and BiP, along with phosphorylation status of p38 MAPK. Note activation of IRE1α, PERK, and ATF6 in ND1 mutants that is suppressed with doxycycline in a MALSU1‐dependent fashion. (F) Genes downstream of all three arms of the UPR are activated with galactose, and suppressed with doxycycline. Genes from respective pathways include IRE1a‐XBP1s arm (DNAJB9, SEC24D), ATF6 arm (HSPA5), and PERK (DDIT3, PPP1R15A). qPCR data represents the average fold change expression levels from the average of n = 4 technical replicates across n = 2 biological samples.
Whole cell IRE1α oligomerization states under galactose conditions assessed by blue‐native PAGE. BN‐PAGE of IRE1α with whole cell extracts does not capture doxycycline regulation of high‐order oligomers when compared to ER concentrated at the mitochondria (Mito/ER extracts, Fig 4B). Glimepiride (25 μM) and IRE1α inhibitor 4μ8C (20 μM), compounds previously identified to rescue complex I mutants from nutrient deprivation, were used as controls. Note compensatory increase in IRE1α expression and oligomerization in 4μ8C treated cells.
The activation of p38 MAPK is downstream of IRE1α and can be modulated by ERAD and mitochondrial translation. WT and ND1 cells were cultured in galactose media for 48 h and analyzed for the activation of MAPK and UPR signaling as evidenced by phosphorylation of p38 (P‐p38) and expression of XBP1s, respectively. Pharmacological inhibition of IRE1α with 4μ8C (20 μM) and p97 inhibitor ESI (20 μM) indicate P‐p38 and XBP1s are downstream of IRE1α and can be controlled by ERAD. CMT3 (1 μM) does not affect P‐p38 MAPK or XBP1s levels when compared to doxycycline (1 μM) indicating inhibition of mitochondrial translation, not off target effects of tetracyclines, modulates UPR and MAPK signaling.
UPR and MAPK signaling are activated after 48 h of galactose stress in ND1 cybrids, where doxycycline (1 μM) and chemically distinct 7002 (1 μM), but not CMT3 (1 μM) attenuate these responses.
Doxycycline suppresses p38 MAPK phosphorylation depending on MALSU1. Data represents the average ± s.e.m. p38 phosphorylation level normalized to total p38 using western blot densitometry (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistics across glucose, galactose, and doxycycline treated cultures in WT, ND1, and sgMALSU1 cells was calculated using two‐way ANOVA, multiple comparisons.

Doxycycline dose‐dependently inhibits UPR and MAPK signaling in ND1 non‐target control (NTC) cells and cells depleted of MARCH5 under galactose conditions. Note basal levels of XBP1s and P‐p38 are increased in sgMARCH5 cells, indicating enhanced sensitivity to ER stress under these conditions.
ND1 cybrids cultured in galactose conditions for 4 days can be rescued by doxycycline independent of MARCH5 expression status (error bars represent the average ± s.e.m. of n = 8 independent biological replicates across N = 3 independent experiments). Statistics of doxycycline treated and untreated cultures across NTC and sgMARCH5 cells were calculated using two‐way ANOVA, multiple comparisons.

- A, B
Dose‐dependent treatment with p97 specific ERAD inhibitor eeyarestatin I (ESI) shows (A) mitochondrial mutant cells are more dependent on ERAD when compared to (B) WT as indicated by enhanced potency (IC50) of ESI in ND1 cells under galactose‐stress conditions. (B) Note ESI potency is unchanged between glucose and galactose conditions in WT cells (IC50). Data points in (A) and (B) represent the average of n = 2 biological replicates.
- C
Inhibition of ERAD with ESI does not reverse doxycycline‐induced survival (error bars represent the average ± s.e.m. of n = 3 independent biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells with or without ESI was calculated using two‐way ANOVA, multiple comparisons.
- D, E
Dose–response curves of secretion inhibitor brefeldin A (BFA) in WT and ND1 cells under glucose and galactose conditions. Note cells are less sensitive to BFA under galactose conditions (IC50 = 25–33 nM) when compared to glucose (IC50 = 8–17 nM) in WT and ND1 cells, respectively. Data points represent the average of n = 2 biological replicates.
- F
Inhibition of secretion with BFA does not reverse doxycycline‐induced survival (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistical significance (P < 0.05) between doxycycline treated and untreated cells with or without BFA was calculated using two‐way ANOVA, multiple comparisons.
- G
Inhibition of ERAD and secretion induce ER stress and the UPR that can be reversed by doxycycline, evidenced by IRE1α and p38 MAPK phosphorylation and XBP1s expression levels. Note enhanced galactose‐induced expression levels of XBP1s and basal levels of BiP that indicate ESI and BFA are on target.
- H
ER protein cathepsin‐C loading is increased in isolated microsomes (ER) upon mitochondrial mutation, which is attenuated by doxycycline. Whole cell (WC) expression levels are unchanged between WT and ND1 cells. Cycloheximide treatment indicates comparable half‐lives between control and doxycycline treated cultures and isolated microsomes.
- I
Microsomal cathepsin‐C levels that are increased upon ND1 mutation are attenuated by doxycycline, and associated with decreases in BiP and SRP14 in these fractions.
- J
Depletion of MALSU1 in ND1 cells reverses doxycycline regulation of BiP, cathepsin‐C, and SRP14 at the microsome.

Tetracyclines do not alter global translation rates as evidenced by puromycin incorporation assays. ND1 cells cultured for 48 h in glucose, galactose, and galactose supplemented with doxycycline (1 μM) and pulsed with puromycin to assay global translation rates. Note decrease in translation upon glucose starvation that is not further altered with doxycycline (representative of n = 2 independent experiments).
Activation of the integrated stress response with mitochondrial DELE1 is not required for tetracyclines to promote survival in mitochondrial mutant cells. ND1 cybrid cells treated with mitochondrial targeting agents (6 h) activate the integrated stress response as indicated by increases in P‐eif2α and ATF4. Note changes in P‐eif2α and ATF4 seen in NTC cells are suppressed in cells lacking DELE1 (sgDELE1). Cells were cultured under glucose conditions except for where noted (galactose). Doxycycline (1 μM, 100 μM), antimycin A (40 nM), and Oligomycin (1.25 ng/μl).
ND1 cells deprived of glucose for 7 days can be rescued by doxycycline (1 μM) independent of DELE1 expression status (error bars represent the average ± s.e.m. of n = 3 biological replicates). Statistics of doxycycline treated and untreated cultures across NTC and sgDELE1 cells were calculated using two‐way ANOVA, multiple comparisons.
Proposed model of pro‐survival MALSU1‐dependent resolution of ER stress by tetracyclines through attenuation of ER stress and IRE1α signaling.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

