Characterization of a novel aromatic ring-hydroxylating oxygenase, NarA2B2, from thermophilic Hydrogenibacillus sp. strain N12
- PMID: 37819076
- PMCID: PMC10617421
- DOI: 10.1128/aem.00865-23
Characterization of a novel aromatic ring-hydroxylating oxygenase, NarA2B2, from thermophilic Hydrogenibacillus sp. strain N12
Abstract
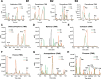
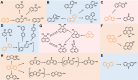
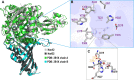
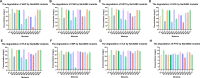
Polycyclic aromatic hydrocarbons (PAHs) are harmful to human health due to their carcinogenic, teratogenic, and mutagenic effects. A thermophilic Hydrogenibacillus sp. strain N12 capable of degrading a variety of PAHs and derivatives was previously isolated. In this study, an aromatic ring-hydroxylating oxygenase, NarA2B2, was identified from strain N12, with substrate specificity including naphthalene, phenanthrene, dibenzothiophene, fluorene, acenaphthene, carbazole, biphenyl, and pyrene. NarA2B2 was proposed to add one or two atoms of molecular oxygen to the substrate and catalyze biphenyl at C-2, 2 or C-3, 4 positions with different characteristics than before. The key catalytic amino acids, H222, H227, and D379, were identified as playing a pivotal role in the formation of the 2-his-1-carboxylate facial triad. Furthermore, we conducted molecular docking and molecular dynamics simulations, notably, D219 enhanced the stability of the iron center by forming two stable hydrogen bonds with H222, while the mutation of F216, T223, and H302 modulated the catalytic activity by altering the pocket's size and shape. Compared to the wild-type (WT) enzyme, the degradation ratios of acenaphthene by F216A, T223A, and H302A had an improvement of 23.08%, 26.87%, and 29.52%, the degradation ratios of naphthalene by T223A and H302A had an improvement of 51.30% and 65.17%, while the degradation ratio of biphenyl by V236A had an improvement of 77.94%. The purified NarA2B2 was oxygen-sensitive when it was incubated with L-ascorbic acid in an anaerobic environment, and its catalytic activity was restored in vitro. These results contribute to a better understanding of the molecular mechanism responsible for PAHs' degradation in thermophilic microorganisms.IMPORTANCE(i) A novel aromatic ring-hydroxylating oxygenase named NarA2B2, capable of degrading multiple polycyclic aromatic hydrocarbons and derivatives, was identified from the thermophilic microorganism Hydrogenibacillus sp. N12. (ii) The degradation characteristics of NarA2B2 were characterized by adding one or two atoms of molecular oxygen to the substrate. Unlike the previous study, NarA2B2 catalyzed biphenyl at C-2, 2 or C-3, 4 positions. (iii) Catalytic sites of NarA2B2 were conserved, and key amino acids F216, D219, H222, T223, H227, V236, F243, Y300, H302, W316, F369, and D379 played pivotal roles in catalysis, as confirmed by protein structure prediction, molecular docking, molecular dynamics simulations, and point mutation.
Keywords: Hydrogenibacillus; aromatic ring-hydroxylating oxygenase; biodegradation; polycyclic aromatic hydrocarbons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Haritash AK, Kaushik CP. 2011. Seasonal and spatial occurrence and distribution of respirable particulate-bound atmospheric polycyclic aromatic hydrocarbons in Hisar city (India) and their potential health-risks. Asian J Water Environ Pollut 8:73–80.
-
- Menzie CA, Potocki BB, Santodonato J. 1992. Exposure to carcinogenic PAHs in the environment. Environ Sci Technol 26:1278–1284. doi: 10.1021/es00031a002 - DOI
-
- Gupta S, Pathak B, Fulekar MH. 2015. Molecular approaches for biodegradation of polycyclic aromatic hydrocarbon compounds: a review. Rev Environ Sci Biotechnol 14:241–269. doi: 10.1007/s11157-014-9353-3 - DOI
-
- Müller R, Antranikian G, Maloney S, Sharp R. 1998. Thermophilic degradation of environmental pollutants, p 155–169. In Advances in biochemical engineering/biotechnology. doi: 10.1007/BFb0102286 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

