Diamide-based screening method for the isolation of improved oxidative stress tolerance phenotypes in Bacillus mutant libraries
- PMID: 37819171
- PMCID: PMC10714788
- DOI: 10.1128/spectrum.01608-23
Diamide-based screening method for the isolation of improved oxidative stress tolerance phenotypes in Bacillus mutant libraries
Abstract
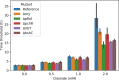
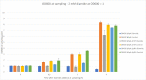
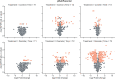
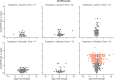
During their life cycle, bacteria are exposed to a range of different stresses that need to be managed appropriately in order to ensure their growth and viability. This applies not only to bacteria in their natural habitats but also to bacteria employed in biotechnological production processes. Oxidative stress is one of these stresses that may originate either from bacterial metabolism or external factors. In biotechnological settings, it is of critical importance that production strains are resistant to oxidative stresses. Accordingly, this also applies to the major industrial cell factory Bacillus subtilis. In the present study, we, therefore, developed a screen for B. subtilis strains with enhanced oxidative stress tolerance. The results show that our approach is feasible and time-, space-, and resource-efficient. We, therefore, anticipate that it will enhance the development of more robust industrial production strains with improved robustness under conditions of oxidative stress.
Keywords: Bacillus subtilis; diamide; disulfide; library screening; mutagenesis.
Conflict of interest statement
J.W., R.S., and M.S. are employees of AB Enzymes GmbH.
Figures



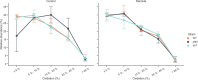
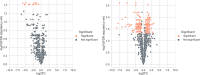
Similar articles
-
Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus.J Bacteriol. 2009 Dec;191(24):7520-30. doi: 10.1128/JB.00937-09. Epub 2009 Oct 16. J Bacteriol. 2009. PMID: 19837798 Free PMC article.
-
The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis.Proteomics. 2010 Sep;10(17):3155-64. doi: 10.1002/pmic.201000230. Proteomics. 2010. PMID: 20652907
-
UbK is Involved in the Resistance of Bacillus Subtilis to Oxidative Stress.Curr Microbiol. 2020 Dec;77(12):4063-4071. doi: 10.1007/s00284-020-02239-1. Epub 2020 Oct 12. Curr Microbiol. 2020. PMID: 33044618
-
Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism.Biochim Biophys Acta. 2004 Nov 11;1694(1-3):299-310. doi: 10.1016/j.bbamcr.2004.02.011. Biochim Biophys Acta. 2004. PMID: 15546673 Review.
-
Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond.World J Microbiol Biotechnol. 2018 Sep 10;34(10):145. doi: 10.1007/s11274-018-2531-7. World J Microbiol Biotechnol. 2018. PMID: 30203131 Review.
Cited by
-
From the outer space to the inner cell: deconvoluting the complexity of Bacillus subtilis disulfide stress responses by redox state and absolute abundance quantification of extracellular, membrane, and cytosolic proteins.Microbiol Spectr. 2024 Apr 2;12(4):e0261623. doi: 10.1128/spectrum.02616-23. Epub 2024 Feb 15. Microbiol Spectr. 2024. PMID: 38358275 Free PMC article.
References
-
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Hènaut A, Hilbert H, Holsappel S, Hosono S, Hullo MF, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee SM, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado RP, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park SH, Parro V, Pohl TM, Portelle D, Porwollik S, Prescott AM, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror SJ, Serror P, Shin BS, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Togoni A, Tosato V, Uchiyama S, Vandebol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa HF, Zumstein E, Yoshikawa H, Danchin A. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi:10.1038/36786 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

