Actin-dependent recruitment of AGO2 to the zonula adherens
- PMID: 37819702
- PMCID: PMC10848941
- DOI: 10.1091/mbc.E22-03-0099-T
Actin-dependent recruitment of AGO2 to the zonula adherens
Abstract
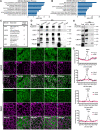
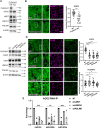
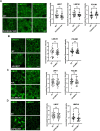
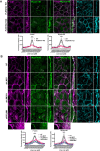
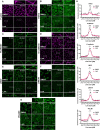
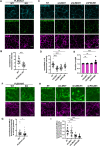
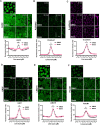
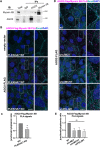
Adherens junctions are cadherin-based structures critical for cellular architecture. E-cadherin junctions in mature epithelial cell monolayers tether to an apical actomyosin ring to form the zonula adherens (ZA). We have previously shown that the adherens junction protein PLEKHA7 associates with and regulates the function of the core RNA interference (RNAi) component AGO2 specifically at the ZA. However, the mechanism mediating AGO2 recruitment to the ZA remained unexplored. Here, we reveal that this ZA-specific recruitment of AGO2 depends on both the structural and tensile integrity of the actomyosin cytoskeleton. We found that depletion of not only PLEKHA7, but also either of the three PLEKHA7-interacting, LIM-domain family proteins, namely LMO7, LIMCH1, and PDLIM1, results in disruption of actomyosin organization and tension, as well as disruption of AGO2 junctional localization and of its miRNA-binding ability. We also show that AGO2 binds Myosin IIB and that PLEKHA7, LMO7, LIMCH1, and PDLIM1 all disrupt interaction of AGO2 with Myosin IIB at the ZA. These results demonstrate that recruitment of AGO2 to the ZA is sensitive to actomyosin perturbations, introducing the concept of mechanosensitive RNAi machinery, with potential implications in tissue remodeling and in disease.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

