AGAMOUS mediates timing of guard cell formation during gynoecium development
- PMID: 37819989
- PMCID: PMC10593234
- DOI: 10.1371/journal.pgen.1011000
AGAMOUS mediates timing of guard cell formation during gynoecium development
Abstract
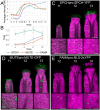
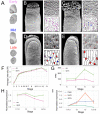
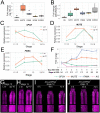
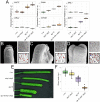
In Arabidopsis thaliana, stomata are composed of two guard cells that control the aperture of a central pore to facilitate gas exchange between the plant and its environment, which is particularly important during photosynthesis. Although leaves are the primary photosynthetic organs of flowering plants, floral organs are also photosynthetically active. In the Brassicaceae, evidence suggests that silique photosynthesis is important for optimal seed oil content. A group of transcription factors containing MADS DNA binding domains is necessary and sufficient to confer floral organ identity. Elegant models, such as the ABCE model of flower development and the floral quartet model, have been instrumental in describing the molecular mechanisms by which these floral organ identity proteins govern flower development. However, we lack a complete understanding of how the floral organ identity genes interact with the underlying leaf development program. Here, we show that the MADS domain transcription factor AGAMOUS (AG) represses stomatal development on the gynoecial valves, so that maturation of stomatal complexes coincides with fertilization. We present evidence that this regulation by AG is mediated by direct transcriptional repression of a master regulator of the stomatal lineage, MUTE, and show data that suggests this interaction is conserved among several members of the Brassicaceae. This work extends our understanding of the mechanisms underlying floral organ formation and provides a framework to decipher the mechanisms that control floral organ photosynthesis.
Copyright: © 2023 Brazel et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors do not have any competing interests.
Figures



References
-
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA Genes Regulate Growth and Pattern Formation during Gynoecium Development in Arabidopsis thaliana. International Journal of Plant Sciences. 2002;163: 17–41. doi: 10.1086/324178 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

