Mass-Activated Droplet Sorting for the Selection of Lysine-Producing Escherichia coli
- PMID: 37820298
- PMCID: PMC11025463
- DOI: 10.1021/acs.analchem.3c03080
Mass-Activated Droplet Sorting for the Selection of Lysine-Producing Escherichia coli
Abstract
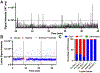
Synthetic biology relies on engineering cells to have desirable properties, such as the production of select chemicals. A bottleneck in engineering methods is often the need to screen and sort variant libraries for potential activity. Droplet microfluidics is a method for high-throughput sample preparation and analysis which has the potential to improve the engineering of cells, but a limitation has been the reliance on fluorescent analysis. Here, we show the ability to select cell variants grown in 20 nL droplets at 0.5 samples/s using mass-activated droplet sorting (MADS), a method for selecting droplets based on the signal intensity measured by electrospray ionization mass spectrometry (ESI-MS). Escherichia coli variants producing lysine were used to evaluate the applicability of MADS for synthetic biology. E. coli were shown to be effectively grown in droplets, and the lysine produced by these cells was detectable using ESI-MS. Sorting of lysine-producing cells based on the MS signal was shown, yielding 96-98% purity for high-producing variants in the selected pool. Using this technique, cells were recovered after screening, enabling downstream validation via phenotyping. The presented method is translatable to whole-cell engineering for biocatalyst production.
Conflict of interest statement
Figures
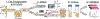
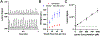

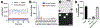
References
-
- Meng D-C; Shi Z-Y; Wu L-P; Zhou Q; Wu Q; Chen J-C; Chen G-Q Production and Characterization of Poly(3-Hydroxypropionate-Co-4-Hydroxybutyrate) with Fully Controllable Structures by Recombinant Escherichia Coli Containing an Engineered Pathway. Metabolic Engineering 2012, 14 (4), 317–324. 10.1016/j.ymben.2012.04.003. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

