Barley MLA3 recognizes the host-specificity effector Pwl2 from Magnaporthe oryzae
- PMID: 37820736
- PMCID: PMC10827324
- DOI: 10.1093/plcell/koad266
Barley MLA3 recognizes the host-specificity effector Pwl2 from Magnaporthe oryzae
Abstract
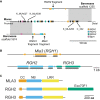
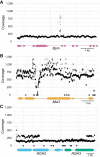
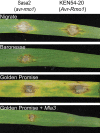
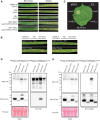
Plant nucleotide-binding leucine-rich repeat (NLRs) immune receptors directly or indirectly recognize pathogen-secreted effector molecules to initiate plant defense. Recognition of multiple pathogens by a single NLR is rare and usually occurs via monitoring for changes to host proteins; few characterized NLRs have been shown to recognize multiple effectors. The barley (Hordeum vulgare) NLR gene Mildew locus a (Mla) has undergone functional diversification, and the proteins encoded by different Mla alleles recognize host-adapted isolates of barley powdery mildew (Blumeria graminis f. sp. hordei [Bgh]). Here, we show that Mla3 also confers resistance to the rice blast fungus Magnaporthe oryzae in a dosage-dependent manner. Using a forward genetic screen, we discovered that the recognized effector from M. oryzae is Pathogenicity toward Weeping Lovegrass 2 (Pwl2), a host range determinant factor that prevents M. oryzae from infecting weeping lovegrass (Eragrostis curvula). Mla3 has therefore convergently evolved the capacity to recognize effectors from diverse pathogens.
Published by Oxford University Press on behalf of American Society of Plant Biologists 2023.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



Similar articles
-
The Magnaporthe oryzae effector Pwl2 alters HIPP43 localization to suppress host immunity.Plant Cell. 2025 Jun 4;37(6):koaf116. doi: 10.1093/plcell/koaf116. Plant Cell. 2025. PMID: 40341381 Free PMC article.
-
Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6486-E6495. doi: 10.1073/pnas.1612947113. Epub 2016 Oct 4. Proc Natl Acad Sci U S A. 2016. PMID: 27702901 Free PMC article.
-
The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold.PLoS Pathog. 2021 Feb 3;17(2):e1009223. doi: 10.1371/journal.ppat.1009223. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33534797 Free PMC article.
-
Specific Resistance of Barley to Powdery Mildew, Its Use and Beyond. A Concise Critical Review.Genes (Basel). 2020 Aug 21;11(9):971. doi: 10.3390/genes11090971. Genes (Basel). 2020. PMID: 32825722 Free PMC article. Review.
-
Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol. 2014 Oct;204(2):273-81. doi: 10.1111/nph.12889. New Phytol. 2014. PMID: 25453131 Review.
Cited by
-
Uncovering the Mechanisms: The Role of Biotrophic Fungi in Activating or Suppressing Plant Defense Responses.J Fungi (Basel). 2024 Sep 5;10(9):635. doi: 10.3390/jof10090635. J Fungi (Basel). 2024. PMID: 39330396 Free PMC article. Review.
-
Adaptive evolution in virulence effectors of the rice blast fungus Pyricularia oryzae.PLoS Pathog. 2023 Sep 11;19(9):e1011294. doi: 10.1371/journal.ppat.1011294. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37695773 Free PMC article.
-
Resurrection of the Plant Immune Receptor Sr50 to Overcome Pathogen Immune Evasion.bioRxiv [Preprint]. 2025 Feb 11:2024.08.07.607039. doi: 10.1101/2024.08.07.607039. bioRxiv. 2025. PMID: 39149390 Free PMC article. Preprint.
-
The resistance awakens: Diversity at the DNA, RNA, and protein levels informs engineering of plant immune receptors from Arabidopsis to crops.Plant Cell. 2025 May 9;37(5):koaf109. doi: 10.1093/plcell/koaf109. Plant Cell. 2025. PMID: 40344182 Free PMC article. Review.
-
The roles of Magnaporthe oryzae avirulence effectors involved in blast resistance/susceptibility.Front Plant Sci. 2024 Oct 9;15:1478159. doi: 10.3389/fpls.2024.1478159. eCollection 2024. Front Plant Sci. 2024. PMID: 39445147 Free PMC article. Review.
References
-
- Ahn H-K, Lin X, Olave-Achury AC, Derevnina L, Contreras MP, Kourelis J, Wu C-H, Kamoun S, Jones JDG. Effector-dependent activation and oligomerization of plant NRC class helper NLRs by sensor NLR immune receptors Rpi-amr3 and Rpi-amr1. EMBO J. 2023:42(5):e111484. 10.15252/embj.2022111484 - DOI - PMC - PubMed
-
- Ashfield T, Redditt T, Russell A, Kessens R, Rodibaugh N, Galloway L, Kang Q, Podicheti R, Innes RW. Evolutionary relationship of disease resistance genes in soybean and Arabidopsis specific for the Pseudomonas syringae effectors AvrB and AvrRpm1. Plant Physiol. 2014:166(1):235–251. 10.1104/pp.114.244715 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- BB/M011216/1/United Kingdom Research and Innovation-Biotechnology and Biological Sciences Research Council Norwich Research Park Doctoral Training Partnership
- BB/P012574/1/Institute Strategic Programme
- CZ.02.1.01/0.0/0.0/16_019/0000827/European Regional Development Fund
- Perry Foundation
- Japan Society for the Promotion of Science 2018 Summer Programme
LinkOut - more resources
Full Text Sources
Research Materials

