Single-cell multi-omics analysis identifies two distinct phenotypes of newly-onset microscopic polyangiitis
- PMID: 37821442
- PMCID: PMC10567716
- DOI: 10.1038/s41467-023-41328-0
Single-cell multi-omics analysis identifies two distinct phenotypes of newly-onset microscopic polyangiitis
Abstract
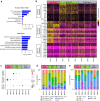
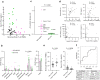
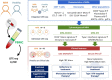
The immunological basis of the clinical heterogeneity in autoimmune vasculitis remains poorly understood. In this study, we conduct single-cell transcriptome analyses on peripheral blood mononuclear cells (PBMCs) from newly-onset patients with microscopic polyangiitis (MPA). Increased proportions of activated CD14+ monocytes and CD14+ monocytes expressing interferon signature genes (ISGs) are distinctive features of MPA. Patient-specific analysis further classifies MPA into two groups. The MPA-MONO group is characterized by a high proportion of activated CD14+ monocytes, which persist before and after immunosuppressive therapy. These patients are clinically defined by increased monocyte ratio in the total PBMC count and have a high relapse rate. The MPA-IFN group is characterized by a high proportion of ISG+ CD14+ monocytes. These patients are clinically defined by high serum interferon-alpha concentrations and show good response to immunosuppressive therapy. Our findings identify the immunological phenotypes of MPA and provide clinical insights for personalized treatment and accurate prognostic prediction.
© 2023. Springer Nature Limited.
Conflict of interest statement
A.K. has received grant support from Chugai Pharmaceutical Co, Ltd. K.N., H.M., S.I., H.K., R.O. and K.H. are employed by Chugai Pharmaceutical Co, Ltd. and K.N., H.M., S.I., H.K., R.O. and K.H. also hold stocks in the company. The remaining authors declare no competing interests.
Figures



References
-
- Hellmich B, et al. EULAR recommendations for the management of ANCA-associated vasculitis: 2022 update. Ann. Rheum. Dis. 2023;16:ard-2022–223764. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

